Identification of specific prognostic markers for lung squamous cell carcinoma based on tumor progression, immune infiltration, and stem index
- PMID: 37841237
- PMCID: PMC10570622
- DOI: 10.3389/fimmu.2023.1236444
Identification of specific prognostic markers for lung squamous cell carcinoma based on tumor progression, immune infiltration, and stem index
Abstract
Introduction: Lung squamous cell carcinoma (LUSC) is a unique subform of nonsmall cell lung cancer (NSCLC). The lack of specific driver genes as therapeutic targets leads to worse prognoses in patients with LUSC, even with chemotherapy, radiotherapy, or immune checkpoint inhibitors. Furthermore, research on the LUSC-specific prognosis genes is lacking. This study aimed to develop a comprehensive LUSC-specific differentially expressed genes (DEGs) signature for prognosis correlated with tumor progression, immune infiltration,and stem index.
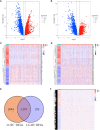
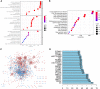
Methods: RNA sequencing data for LUSC and lung adenocarcinoma (LUAD) were extracted from The Cancer Genome Atlas (TCGA) data portal, and DEGs analyses were conducted in TCGA-LUSC and TCGA-LUAD cohorts to identify specific DEGs associated with LUSC. Functional analysis and protein-protein interaction network were performed to annotate the roles of LUSC-specific DEGs and select the top 100 LUSC-specific DEGs. Univariate Cox regression and least absolute shrinkage and selection operator regression analyses were performed to select prognosis-related DEGs.
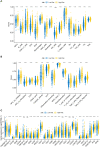
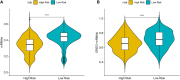
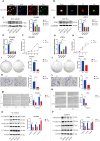
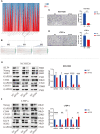
Results: Overall, 1,604 LUSC-specific DEGs were obtained, and a validated seven-gene signature was constructed comprising FGG, C3, FGA, JUN, CST3, CPSF4, and HIST1H2BH. FGG, C3, FGA, JUN, and CST3 were correlated with poor LUSC prognosis, whereas CPSF4 and HIST1H2BH were potential positive prognosis markers in patients with LUSC. Receiver operating characteristic analysis further confirmed that the genetic profile could accurately estimate the overall survival of LUSC patients. Analysis of immune infiltration demonstrated that the high risk (HR) LUSC patients exhibited accelerated tumor infiltration, relative to low risk (LR) LUSC patients. Molecular expressions of immune checkpoint genes differed significantly between the HR and LR cohorts. A ceRNA network containing 19 lncRNAs, 50 miRNAs, and 7 prognostic DEGs was constructed to demonstrate the prognostic value of novel biomarkers of LUSC-specific DEGs based on tumor progression, stemindex, and immune infiltration. In vitro experimental models confirmed that LUSC-specific DEG FGG expression was significantly higher in tumor cells and correlated with immune tumor progression, immune infiltration, and stem index. In vitro experimental models confirmed that LUSC-specific DEG FGG expression was significantly higher in tumor cells and correlated with immune tumor progression, immune infiltration, and stem index.
Conclusion: Our study demonstrated the potential clinical implication of the 7- DEGs signature for prognosis prediction of LUSC patients based on tumor progression, immune infiltration, and stem index. And the FGG could be an independent prognostic biomarker of LUSC promoting cell proliferation, migration, invasion, THP-1 cell infiltration, and stem cell maintenance.
Keywords: LUSC; biomarker; cancer stem cell; prognosis; tumor microenvironment.
Copyright © 2023 Wu, Ma, Duan, Zhang, Li, Yu, Zhang, Liu and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

