Identification of immune-related biomarkers in peripheral blood of schizophrenia using bioinformatic methods and machine learning algorithms
- PMID: 37841288
- PMCID: PMC10568181
- DOI: 10.3389/fncel.2023.1256184
Identification of immune-related biomarkers in peripheral blood of schizophrenia using bioinformatic methods and machine learning algorithms
Abstract
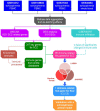
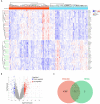
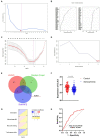
Schizophrenia is a group of severe neurodevelopmental disorders. Identification of peripheral diagnostic biomarkers is an effective approach to improving diagnosis of schizophrenia. In this study, four datasets of schizophrenia patients' blood or serum samples were downloaded from the GEO database and merged and de-batched for the analyses of differentially expressed genes (DEGs) and weighted gene co-expression network analysis (WCGNA). The WGCNA analysis showed that the cyan module, among 9 modules, was significantly related to schizophrenia, which subsequently yielded 317 schizophrenia-related key genes by comparing with the DEGs. The enrichment analyses on these key genes indicated a strong correlation with immune-related processes. The CIBERSORT algorithm was adopted to analyze immune cell infiltration, which revealed differences in eosinophils, M0 macrophages, resting mast cells, and gamma delta T cells. Furthermore, by comparing with the immune genes obtained from online databases, 95 immune-related key genes for schizophrenia were screened out. Moreover, machine learning algorithms including Random Forest, LASSO, and SVM-RFE were used to further screen immune-related hub genes of schizophrenia. Finally, CLIC3 was found as an immune-related hub gene of schizophrenia by the three machine learning algorithms. A schizophrenia rat model was established to validate CLIC3 expression and found that CLIC3 levels were reduced in the model rat plasma and brains in a brain-regional dependent manner, but can be reversed by an antipsychotic drug risperidone. In conclusion, using various bioinformatic and biological methods, this study found an immune-related hub gene of schizophrenia - CLIC3 that might be a potential diagnostic biomarker and therapeutic target for schizophrenia.
Keywords: CIBERSORT; CLIC3; LASSO; WGCNA; peripheral immune-related biomarkers; random forest; schizophrenia; support vector machine.
Copyright © 2023 Zhu, Wang, Yu, Weng, Han, Liu, Tang and Pan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Identification of Immune-Related Biomarkers of Schizophrenia in the Central Nervous System Using Bioinformatic Methods and Machine Learning Algorithms.Mol Neurobiol. 2025 Mar;62(3):3226-3243. doi: 10.1007/s12035-024-04461-5. Epub 2024 Sep 7. Mol Neurobiol. 2025. PMID: 39243324
-
Identification of novel biomarkers and immune infiltration characteristics of ischemic stroke based on comprehensive bioinformatic analysis and machine learning.Biochem Biophys Rep. 2023 Dec 7;37:101595. doi: 10.1016/j.bbrep.2023.101595. eCollection 2024 Mar. Biochem Biophys Rep. 2023. PMID: 38371524 Free PMC article.
-
Six potential biomarkers in septic shock: a deep bioinformatics and prospective observational study.Front Immunol. 2023 Jun 8;14:1184700. doi: 10.3389/fimmu.2023.1184700. eCollection 2023. Front Immunol. 2023. PMID: 37359526 Free PMC article.
-
Utilizing machine learning algorithms to identify biomarkers associated with diabetic nephropathy: A review.Medicine (Baltimore). 2024 Feb 23;103(8):e37235. doi: 10.1097/MD.0000000000037235. Medicine (Baltimore). 2024. PMID: 38394492 Free PMC article. Review.
-
Identification of biomarkers associated with pediatric asthma using machine learning algorithms: A review.Medicine (Baltimore). 2023 Nov 24;102(47):e36070. doi: 10.1097/MD.0000000000036070. Medicine (Baltimore). 2023. PMID: 38013370 Free PMC article. Review.
Cited by
-
Artificial Intelligence in Psychiatry: A Review of Biological and Behavioral Data Analyses.Diagnostics (Basel). 2025 Feb 11;15(4):434. doi: 10.3390/diagnostics15040434. Diagnostics (Basel). 2025. PMID: 40002587 Free PMC article. Review.
-
Application of machine learning for the analysis of peripheral blood biomarkers in oral mucosal diseases: a cross-sectional study.BMC Oral Health. 2025 May 10;25(1):703. doi: 10.1186/s12903-025-06095-y. BMC Oral Health. 2025. PMID: 40348983 Free PMC article.
-
Biomarker discovery using machine learning in the psychosis spectrum.Biomark Neuropsychiatry. 2024 Dec;11:100107. doi: 10.1016/j.bionps.2024.100107. Epub 2024 Aug 26. Biomark Neuropsychiatry. 2024. PMID: 39687745 Free PMC article.
-
White Blood Cell Proportions Are Associated With Response to Psychosocial Therapy in Young People at Ultra-High Risk for Psychosis.Biol Psychiatry Glob Open Sci. 2025 Jun 3;5(5):100546. doi: 10.1016/j.bpsgos.2025.100546. eCollection 2025 Sep. Biol Psychiatry Glob Open Sci. 2025. PMID: 40697486 Free PMC article.
-
Exploring the Intersection of Schizophrenia, Machine Learning, and Genomics: Scoping Review.JMIR Bioinform Biotechnol. 2024 Nov 15;5:e62752. doi: 10.2196/62752. JMIR Bioinform Biotechnol. 2024. PMID: 39546776 Free PMC article.
References
LinkOut - more resources
Full Text Sources

