Genome-wide identification of the TCP gene family in Chrysanthemum lavandulifolium and its homologs expression patterns during flower development in different Chrysanthemum species
- PMID: 37841609
- PMCID: PMC10570465
- DOI: 10.3389/fpls.2023.1276123
Genome-wide identification of the TCP gene family in Chrysanthemum lavandulifolium and its homologs expression patterns during flower development in different Chrysanthemum species
Abstract
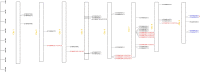
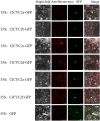
TCP proteins, part of the transcription factors specific to plants, are recognized for their involvement in various aspects of plant growth and development. Nevertheless, a thorough investigation of TCPs in Chrysanthemum lavandulifolium, a prominent ancestral species of cultivated chrysanthemum and an excellent model material for investigating ray floret (RF) and disc floret (DF) development in Chrysanthemum, remains unexplored yet. Herein, a comprehensive study was performed to analyze the genome-wide distribution of TCPs in C. lavandulifolium. In total, 39 TCPs in C. lavandulifolium were identified, showing uneven distribution on 8 chromosomes. Phylogenetic and gene structural analyses revealed that ClTCPs were grouped into classes I and II. The class II genes were subdivided into two subclades, the CIN and CYC/TB1 subclades, with members of each clade having similar conserved motifs and gene structures. Four CIN subclade genes (ClTCP24, ClTCP25, ClTCP26, and ClTCP27) contained the potential miR319 target sites. Promoter analysis revealed that ClTCPs had numerous cis-regulatory elements associated with phytohormone responses, stress responses, and plant growth/development. The expression patterns of ClTCPs during capitulum development and in two different florets were determined using RNA-seq and qRT-PCR. The expression levels of TCPs varied in six development stages of capitula; 25 out of the 36 TCPs genes were specifically expressed in flowers. Additionally, we identified six key ClCYC2 genes, which belong to the class II TCP subclade, with markedly upregulated expression in RFs compared with DFs, and these genes exhibited similar expression patterns in the two florets of Chrysanthemum species. It is speculated that they may be responsible for RFs and DFs development. Subcellular localization and transactivation activity analyses of six candidate genes demonstrated that all of them were localized in the nucleus, while three exhibited self-activation activities. This research provided a better understanding of TCPs in C. lavandulifolium and laid a foundation for unraveling the mechanism by which important TCPs involved in the capitulum development.
Keywords: Chrysanthemum; TCP gene family; capitulum development; expression analysis; ray and disc floret.
Copyright © 2023 Wu, Li, Wen, Zhang and Dai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
The chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse capitulum types.Hortic Res. 2022 Jan 18;9:uhab022. doi: 10.1093/hr/uhab022. Online ahead of print. Hortic Res. 2022. PMID: 35039834 Free PMC article.
-
Genome-wide identification of the MIKCc-type MADS-box gene family in Chrysanthemum lavandulifolium reveals their roles in the capitulum development.Front Plant Sci. 2023 Mar 23;14:1153490. doi: 10.3389/fpls.2023.1153490. eCollection 2023. Front Plant Sci. 2023. PMID: 37035079 Free PMC article.
-
Identification and Characterization of CYC-Like Genes in Regulation of Ray Floret Development in Chrysanthemum morifolium.Front Plant Sci. 2016 Nov 7;7:1633. doi: 10.3389/fpls.2016.01633. eCollection 2016. Front Plant Sci. 2016. PMID: 27872631 Free PMC article.
-
The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development.Plant Signal Behav. 2015;10(7):e1044192. doi: 10.1080/15592324.2015.1044192. Plant Signal Behav. 2015. PMID: 26039357 Free PMC article. Review.
-
Genomic survey of TCP transcription factors in plants: Phylogenomics, evolution and their biology.Front Genet. 2022 Nov 9;13:1060546. doi: 10.3389/fgene.2022.1060546. eCollection 2022. Front Genet. 2022. PMID: 36437962 Free PMC article. Review.
Cited by
-
Genome-wide identification and expression analysis of the TCP transcription factor family and its response to abiotic stress in rapeseed (Brassica napus L.).3 Biotech. 2025 May;15(5):119. doi: 10.1007/s13205-025-04273-x. Epub 2025 Apr 7. 3 Biotech. 2025. PMID: 40201755 Free PMC article.
-
Identification and expression responses of TCP gene family in Opisthopappus taihangensis under abiotic stress.Front Plant Sci. 2025 Mar 6;16:1499244. doi: 10.3389/fpls.2025.1499244. eCollection 2025. Front Plant Sci. 2025. PMID: 40115945 Free PMC article.
-
ECE-CYC1 Transcription Factor CmCYC1a May Interact with CmCYC2 in Regulating Flower Symmetry and Stamen Development in Chrysanthemum morifolium.Genes (Basel). 2025 Jan 26;16(2):152. doi: 10.3390/genes16020152. Genes (Basel). 2025. PMID: 40004481 Free PMC article.
-
Genome-wide identification and characterization of TCP gene family in Dendrobium nobile and their role in perianth development.Front Plant Sci. 2024 Feb 5;15:1352119. doi: 10.3389/fpls.2024.1352119. eCollection 2024. Front Plant Sci. 2024. PMID: 38375086 Free PMC article.
References
-
- Broholm S. K., Taehtiharju S., Laitinen R., Albert V. A., Teeri T. H., Elomaa P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. U. S. A. 105 (26), 9117–9122. doi: 10.1073/pnas.0801359105 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

