Macrophage RIPK3 triggers inflammation and cell death via the XBP1-Foxo1 axis in liver ischaemia-reperfusion injury
- PMID: 37841640
- PMCID: PMC10568422
- DOI: 10.1016/j.jhepr.2023.100879
Macrophage RIPK3 triggers inflammation and cell death via the XBP1-Foxo1 axis in liver ischaemia-reperfusion injury
Abstract
Background & aims: Receptor-interacting serine/threonine-protein kinase 3 (RIPK3) is a central player in triggering necroptotic cell death. However, whether macrophage RIPK3 may regulate NOD1-dependent inflammation and calcineurin/transient receptor potential cation channel subfamily M member 7 (TRPM7)-induced hepatocyte death in oxidative stress-induced liver inflammatory injury remains elusive.
Methods: A mouse model of hepatic ischaemia-reperfusion (IR) injury, the primary hepatocytes, and bone marrow-derived macrophages were used in the myeloid-specific RIPK3 knockout (RIPK3M-KO) and RIPK3-proficient (RIPK3FL/FL) mice.
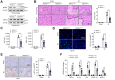
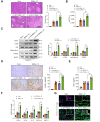
Results: RIPK3M-KO diminished IR stress-induced liver damage with reduced serum alanine aminotransferase/aspartate aminotransferase levels, macrophage/neutrophil infiltration, and pro-inflammatory mediators compared with the RIPK3FL/FL controls. IR stress activated RIPK3, inositol-requiring transmembrane kinase/endoribonuclease 1α (IRE1α), x-box binding protein 1 (XBP1), nucleotide-binding oligomerisation domain-containing protein 1 (NOD1), NF-κB, forkhead box O1 (Foxo1), calcineurin A, and TRPM7 in ischaemic livers. Conversely, RIPK3M-KO depressed IRE1α, XBP1, NOD1, calcineurin A, and TRPM7 activation with reduced serum tumour necrosis factor α (TNF-α) levels. Moreover, Foxo1M-KO alleviated IR-induced liver injury with reduced NOD1 and TRPM7 expression. Interestingly, chromatin immunoprecipitation coupled with massively parallel sequencing revealed that macrophage Foxo1 colocalised with XBP1 and activated its target gene Zc3h15 (zinc finger CCCH domain-containing protein 15). Activating macrophage XBP1 enhanced Zc3h15, NOD1, and NF-κB activity. However, disruption of macrophage Zc3h15 inhibited NOD1 and hepatocyte calcineurin/TRPM7 activation, with reduced reactive oxygen species production and lactate dehydrogenase release after macrophage/hepatocyte coculture. Furthermore, adoptive transfer of Zc3h15-expressing macrophages in RIPK3M-KO mice augmented IR-triggered liver inflammation and cell death.
Conclusions: Macrophage RIPK3 activates the IRE1α-XBP1 pathway and Foxo1 signalling in IR-stress livers. The XBP1-Foxo1 interaction is essential for modulating target gene Zc3h15 function, which is crucial for the control of NOD1 and calcineurin-mediated TRPM7 activation. XBP1 functions as a transcriptional coactivator of Foxo1 in regulating NOD1-driven liver inflammation and calcineurin/TRPM7-induced cell death. Our findings underscore a novel role of macrophage RIPK3 in stress-induced liver inflammation and cell death, implying the potential therapeutic targets in liver inflammatory diseases.
Impact and implications: Macrophage RIPK3 promotes NOD1-dependent inflammation and calcineurin/TRPM7-induced cell death cascade by triggering the XBP1-Foxo1 axis and its target gene Zc3h15, which is crucial for activating NOD1 and calcineurin/TRPM7 function, implying the potential therapeutic targets in stress-induced liver inflammatory injury.
Keywords: ER stress; Foxo1; IRE1α; Innate immunity; Liver inflammation; Necroptosis; Reactive oxygen species; XBP1.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures







Similar articles
-
Novel role of macrophage TXNIP-mediated CYLD-NRF2-OASL1 axis in stress-induced liver inflammation and cell death.JHEP Rep. 2022 Jul 8;4(9):100532. doi: 10.1016/j.jhepr.2022.100532. eCollection 2022 Sep. JHEP Rep. 2022. PMID: 36035360 Free PMC article.
-
Macrophage Notch1 inhibits TAK1 function and RIPK3-mediated hepatocyte necroptosis through activation of β-catenin signaling in liver ischemia and reperfusion injury.Cell Commun Signal. 2022 Sep 16;20(1):144. doi: 10.1186/s12964-022-00901-8. Cell Commun Signal. 2022. PMID: 36114543 Free PMC article.
-
Macrophage Dvl2 deficiency promotes NOD1-Driven pyroptosis and exacerbates inflammatory liver injury.Redox Biol. 2025 Feb;79:103455. doi: 10.1016/j.redox.2024.103455. Epub 2024 Dec 4. Redox Biol. 2025. PMID: 39644526 Free PMC article.
-
Decoding cell death signals in liver inflammation.J Hepatol. 2013 Sep;59(3):583-94. doi: 10.1016/j.jhep.2013.03.033. Epub 2013 Apr 6. J Hepatol. 2013. PMID: 23567086 Review.
-
Forkhead Box O1 Regulates Macrophage Polarization Following Staphylococcus aureus Infection: Experimental Murine Data and Review of the Literature.Clin Rev Allergy Immunol. 2016 Dec;51(3):353-369. doi: 10.1007/s12016-016-8531-1. Clin Rev Allergy Immunol. 2016. PMID: 26924010 Review.
Cited by
-
XBP1 Facilitating NF-κB-p65 Nuclear Translocation Promotes Macrophage-Originated Sterile Inflammation Via Regulating MT2 Transcription in the Ischemia/Reperfusion Liver.Cell Mol Gastroenterol Hepatol. 2024;18(6):101402. doi: 10.1016/j.jcmgh.2024.101402. Epub 2024 Sep 12. Cell Mol Gastroenterol Hepatol. 2024. PMID: 39271015 Free PMC article.
-
Single-Cell RNA-Seq Uncovers Robust Glial Cell Transcriptional Changes in Methamphetamine-Administered Mice.Int J Mol Sci. 2025 Jan 14;26(2):649. doi: 10.3390/ijms26020649. Int J Mol Sci. 2025. PMID: 39859365 Free PMC article.
-
Corticosterone effects induced by stress and immunity and inflammation: mechanisms of communication.Front Endocrinol (Lausanne). 2025 Mar 20;16:1448750. doi: 10.3389/fendo.2025.1448750. eCollection 2025. Front Endocrinol (Lausanne). 2025. PMID: 40182637 Free PMC article. Review.
-
ZC3H15 suppression ameliorates bone cancer pain through inhibiting neuronal oxidative stress and microglial inflammation.Neoplasia. 2025 Mar;61:101123. doi: 10.1016/j.neo.2025.101123. Epub 2025 Feb 4. Neoplasia. 2025. PMID: 39908779 Free PMC article.
-
Association of RIPK1 and RIPK2 Gene Polymorphisms with Rheumatoid Arthritis in a Chinese Han Population.Appl Clin Genet. 2024 Oct 19;17:159-169. doi: 10.2147/TACG.S472418. eCollection 2024. Appl Clin Genet. 2024. PMID: 39444708 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

