This is a preprint.
Pathogenic variants in SMARCA1 cause an X-linked neurodevelopmental disorder modulated by NURF complex composition
- PMID: 37841849
- PMCID: PMC10571636
- DOI: 10.21203/rs.3.rs-3317938/v1
Pathogenic variants in SMARCA1 cause an X-linked neurodevelopmental disorder modulated by NURF complex composition
Update in
-
Pathogenic variants in SMARCA1 cause an X-linked neurodevelopmental disorder modulated by NURF complex composition.Nat Commun. 2025 Nov 10;16(1):9875. doi: 10.1038/s41467-025-64838-5. Nat Commun. 2025. PMID: 41213919 Free PMC article.
Abstract
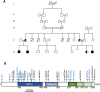
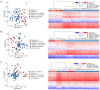
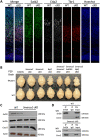
Pathogenic variants in ATP-dependent chromatin remodeling proteins are a recurrent cause of neurodevelopmental disorders (NDDs). The NURF complex consists of BPTF and either the SNF2H (SMARCA5) or SNF2L (SMARCA1) ISWI-chromatin remodeling enzyme. Pathogenic variants in BPTF and SMARCA5 were previously implicated in NDDs. Here, we describe 40 individuals from 30 families with de novo or maternally inherited pathogenic variants in SMARCA1. This novel NDD was associated with mild to severe ID/DD, delayed or regressive speech development, and some recurrent facial dysmorphisms. Individuals carrying SMARCA1 loss-of-function variants exhibited a mild genome-wide DNA methylation profile and a high penetrance of macrocephaly. Genetic dissection of the NURF complex using Smarca1, Smarca5, and Bptfsingle and double mouse knockouts revealed the importance of NURF composition and dosage for proper forebrain development. Finally, we propose that genetic alterations affecting different NURF components result in a NDD with a broad clinical spectrum.
Keywords: NURF complex; SMARCA1; brain overgrowth; epigenetics; exome sequencing.
Conflict of interest statement
KGM and MJGS are employees of GeneDX, LLC. All remaining authors declare no competing financial interests.
Figures



References
-
- Clapier C. R., Cairns B. R., The biology of chromatin remodeling complexes. Annu Rev Biochem 78, 273–304 (2009). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

