This is a preprint.
Dual blockade of BRD4 and ATR/WEE1 pathways exploits ARID1A loss in clear cell ovarian cancer
- PMID: 37841875
- PMCID: PMC10571599
- DOI: 10.21203/rs.3.rs-3314138/v1
Dual blockade of BRD4 and ATR/WEE1 pathways exploits ARID1A loss in clear cell ovarian cancer
Abstract
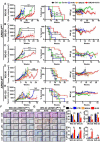
ARID1A, an epigenetic tumor suppressor, is the most common gene mutation in clear-cell ovarian cancers (CCOCs). CCOCs are often resistant to standard chemotherapy and lack effective therapies. We hypothesized that ARID1A loss would increase CCOC cell dependency on chromatin remodeling and DNA repair pathways for survival. We demonstrate that combining BRD4 inhibitor (BRD4i) with DNA damage response inhibitors (ATR or WEE1 inhibitors; e.g. BRD4i-ATRi) was synergistic at low doses leading to decreased survival, and colony formation in CCOC in an ARID1A dependent manner. BRD4i-ATRi caused significant tumor regression and increased overall survival in ARID1AMUT but not ARID1AWT patient-derived xenografts. Combination BRD4i-ATRi significantly increased γH2AX, and decreased RAD51 foci and BRCA1 expression, suggesting decreased ability to repair DNA double-strand-breaks (DSBs) by homologous-recombination in ARID1AMUT cells, and these effects were greater than monotherapies. These studies demonstrate BRD4i-ATRi is an effective treatment strategy that capitalizes on synthetic lethality with ARID1A loss in CCOC.
Keywords: ARID1A; ATR; BRD4; DNA damage response; WEE1; clear cell ovarian cancer.
Conflict of interest statement
Conflict of Interest F.S. serves on scientific advisory boards for AstraZeneca, GSK and Zentalis Pharmaceuticals. She has received institutional research funding from AstraZeneca, Repare Therapeutics, Instill Bio and Sierra Oncology. G.B.M. receives support or acts as a consultant for Amphista, Astex, AstraZeneca, BlueDot, Chrysallis Biotechnology, Ellipses Pharma, GSK, ImmunoMET, Infinity, Ionis, Leapfrog Bio, Lilly, Medacorp, Nanostring, Nuvectis, PDX Pharmaceuticals, Qureator, Roche, Rybodyne, Signalchem Lifesciences, Tarveda, Turbine, and Zentalis Pharmaceuticals has stock options with Bluedot, Biodyne, Catena Pharmaceuticals, ImmunoMet, Nuvectis, RyboDyne, SignalChem, Tarveda, and Turbine, has transferred technology to Myriad and Nanostring and receives sponsored research support from AstraZeneca Nanostring and Zentalis. E.J.B. serves on the scientific advisory board of Atrin Pharmaceuticals and has been an advisor for Sierra Oncology. R.D. serves on the scientific advisory board of Repare Therapeutics and Siamab Therapeutics and advises Mersana Therapeutics and nVision Medical.
Figures






References
-
- Oliver K. E. et al. An evaluation of progression free survival and overall survival of ovarian cancer patients with clear cell carcinoma versus serous carcinoma treated with platinum therapy: An NRG Oncology/Gynecologic Oncology Group experience. Gynecol Oncol 147, 243–249, doi: 10.1016/j.ygyno.2017.08.004 (2017). - DOI - PMC - PubMed
-
- Sugiyama T. et al. Clinical characteristics of clear cell carcinoma of the ovary: a distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Cancer 88, 2584–2589 (2000). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

