Sociodemographic distributions and molecular characterization of colonized Enterococcus faecium isolates from locality hospitals in Khartoum, Sudan
- PMID: 37842047
- PMCID: PMC10573385
- DOI: 10.7717/peerj.16169
Sociodemographic distributions and molecular characterization of colonized Enterococcus faecium isolates from locality hospitals in Khartoum, Sudan
Abstract
Background: Enterococcus faecium is an opportunistic pathogen of humans with diverse hosts, encompassing animals as well as human beings. In the past twenty years, there has been a rise in the instances of nosocomial infections that are linked to antibiotic-resistant Enterococcus faecium. The acquisition of diverse antimicrobial resistance factors has driven the global development of robust and convergent adaptive mechanisms within the healthcare environment. The presence of microorganisms in hospitalized and non-hospitalized patient populations has been significantly aided by the facilitation of various perturbations within their respective microbiomes.
Objective: This study aimed to determine the antimicrobial profile, demographic and clinical characteristics, along with the detection of virulence encoding genes, and to find out the clonal genetic relationship among colonized E. faecium strains.
Methodology: A hospital-based cross-sectional study was carried out between October 2018 and March 2020 at four Khartoum locality hospitals in Sudan. The study comprised a total of 108 strains of E. faecium isolated from patients admitted to four locality hospitals in Khartoum. A self-structured questionnaire was used to gather information on sociodemographic traits. Data were analyzed using chi-square test. In all cases, P value ≤ 0.05 with a corresponding 95% confidence interval was considered statistically significant. Moreover, enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) was utilized to assess the prevalence of clonal relationships, and the gel was analyzed using CLIQS software.
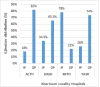
Results: In this study, the isolation rate of colonized E. faecium strains was 108/170 (63.5%). The colonization of E. faecium and its association with various sociodemographic and clinical features was examined. 73 (67.6%) of patients had multidrug-resistant (MDR), and 22 (20.4%) had extensively drug-resistant (XDR), 73 (67.6%) of patients engaged in self-medication practices. Eighty patients (74.1%) were non-adherence to prescribed antibiotics, while 70 (64.8%) patients reported recent antibiotic usage within the 3 months. The present study suggests that demographic factors may not be significantly associated with the incidence of E. faecium infection except for patients who had a prior history of antibiotic use (P ≤ 0.005). The analysis of virulence genes showed a high prevalence of asa1 gene (22.2%) among strains. In ERIC-PCR the genetic relatedness of E. faecium showed seven identical clusters (A-G) with 100% genetic similarity. This implies clonal propagation in hospitals and communities.
Conclusion: This study found that the incidence of E. faecium isolated from locality hospitals in Khartoum was likely due to the spread of E. faecium clones, thereby highlighting the need for intensifying infection control measures to prevent the spreading of nosocomial infection.
Keywords: Antibiotic resistance pattern; Clonal relationship; ERIC-PCR; Enterococcus; Enterococcus faecium; Khartoum locality hospitals; Sociodemographic distribution; Sudan; Vancomycin resistant Enterococcus faecium; Virulence encoding gene.
©2023 Siddig et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


References
-
- Alkhuzaei AMJB, Salama RE, Eljak IEI, Chehab MA, Selim NA. Perceptions and practice of physicians and pharmacists regarding antibiotic misuse at primary health centers in Qatar: a cross-sectional study. Journal of Taibah University Medical Sciences. 2018;13(1):77–82. doi: 10.1016/j.jtumed.2017.09.001. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

