Identification of a novel intermittent hypoxia-related prognostic lncRNA signature and the ceRNA of lncRNA GSEC/miR-873-3p/EGLN3 regulatory axis in lung adenocarcinoma
- PMID: 37842058
- PMCID: PMC10573295
- DOI: 10.7717/peerj.16242
Identification of a novel intermittent hypoxia-related prognostic lncRNA signature and the ceRNA of lncRNA GSEC/miR-873-3p/EGLN3 regulatory axis in lung adenocarcinoma
Abstract
Background: Lung adenocarcinoma (LUAD) is still the most prevalent type of respiratory cancer. Intermittent hypoxia can increase the mortality and morbidity associated with lung cancer. Long non-coding RNAs (lncRNAs) are crucial in lung adenocarcinoma. However, the effects of intermittent hypoxia-related long non-coding RNAs (IHRLs) on lung adenocarcinoma are still unknown.
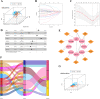
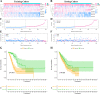
Method: In the current research, eight IHRLs were selected to create a prognostic model. The risk score of the prognostic model was evaluated using multivariate and univariate analyses, and its accuracy and reliability were validated using a nomogram and ROC. Additionally, we investigated the relationships between IHRLs and the immune microenvironment.
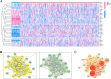
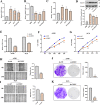
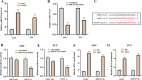
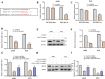
Result: Our analysis identified GSEC, AC099850.3, and AL391001.1 as risk lncRNAs, while AC010615.2, AC010654.1, AL513550.1, LINC00996, and LINC01150 were categorized as protective lncRNAs. We observed variances in the expression of seven immune cells and 15 immune-correlated pathways between the two risk groups. Furthermore, our results confirmed the ceRNA network associated with the intermittent hypoxia-related lncRNA GSEC/miR-873-3p/EGLN3 regulatory pathway. GSEC showed pronounced expression in lung adenocarcinoma tissues and specific cell lines, and its inhibition resulted in reduced proliferation and migration in A549 and PC9 cells. Intriguingly, GSEC manifested oncogenic properties by sponging miR-873-3p and demonstrated a tendency to modulate EGLN3 expression favorably.
Conclusion: GSEC acts as an oncogenic lncRNA by interacting with miR-873-3p, modulating EGLN3 expression. This observation underscores the potential of GSEC as a diagnostic and therapeutic target for LUAD.
Keywords: EGLN3; GSEC; Immune microenvironment; Intermittent hypoxia; Lung adenocarcinoma; MiR-873-3p.
©2023 Liu et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures






Similar articles
-
A novel cuproptosis-related prognostic lncRNA signature and lncRNA MIR31HG/miR-193a-3p/TNFRSF21 regulatory axis in lung adenocarcinoma.Front Oncol. 2022 Jul 22;12:927706. doi: 10.3389/fonc.2022.927706. eCollection 2022. Front Oncol. 2022. PMID: 35936736 Free PMC article.
-
A disulfidptosis-related lncRNA signature for predicting prognosis and evaluating the tumor immune microenvironment of lung adenocarcinoma.Sci Rep. 2024 Feb 26;14(1):4621. doi: 10.1038/s41598-024-55201-7. Sci Rep. 2024. PMID: 38409243 Free PMC article.
-
LncRNA GSEC promotes the proliferation, migration and invasion by sponging miR-588/ EIF5A2 axis in osteosarcoma.Biochem Biophys Res Commun. 2020 Nov 5;532(2):300-307. doi: 10.1016/j.bbrc.2020.08.056. Epub 2020 Aug 29. Biochem Biophys Res Commun. 2020. PMID: 32868080
-
Comprehensive analysis of GSEC/miR-101-3p/SNX16/PAPOLG axis in hepatocellular carcinoma.PLoS One. 2022 Apr 28;17(4):e0267117. doi: 10.1371/journal.pone.0267117. eCollection 2022. PLoS One. 2022. PMID: 35482720 Free PMC article.
-
The role of lncRNAs in intermittent hypoxia and sleep Apnea: A review of experimental and clinical evidence.Sleep Med. 2024 Jan;113:188-197. doi: 10.1016/j.sleep.2023.11.014. Epub 2023 Nov 14. Sleep Med. 2024. PMID: 38043330 Review.
Cited by
-
Ferroptosis-related LncRNAs in diseases.BMC Biol. 2025 Jun 6;23(1):158. doi: 10.1186/s12915-025-02268-x. BMC Biol. 2025. PMID: 40481573 Free PMC article. Review.
-
Construction of a cuproptosis-tricarboxylic acid cycle-associated lncRNA model to predict the prognosis of non-small cell lung cancer.Transl Cancer Res. 2024 Dec 31;13(12):6807-6824. doi: 10.21037/tcr-24-660. Epub 2024 Dec 27. Transl Cancer Res. 2024. PMID: 39816567 Free PMC article.
-
AC099850.3 promotes HBV-HCC cell proliferation and invasion through regulating CD276: a novel strategy for sorafenib and immune checkpoint combination therapy.J Transl Med. 2024 Aug 31;22(1):809. doi: 10.1186/s12967-024-05576-y. J Transl Med. 2024. PMID: 39217342 Free PMC article.
References
-
- Chen L, Qiu CH, Chen Y, Wang Y, Zhao JJ, Zhang M. LncRNA SNHG16 drives proliferation, migration, and invasion of lung cancer cell through modulation of miR-520/VEGF axis. European Review for Medical and Pharmacological Sciences. 2020;24(18):9522–9531. doi: 10.26355/eurrev_202009_23037. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

