Multi-omics data integration reveals the complexity and diversity of host factors associated with influenza virus infection
- PMID: 37842064
- PMCID: PMC10569165
- DOI: 10.7717/peerj.16194
Multi-omics data integration reveals the complexity and diversity of host factors associated with influenza virus infection
Abstract
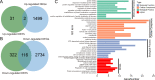
Influenza viruses pose a significant and ongoing threat to human health. Many host factors have been identified to be associated with influenza virus infection. However, there is currently a lack of an integrated resource for these host factors. This study integrated human genes and proteins associated with influenza virus infections for 14 subtypes of influenza A viruses, as well as influenza B and C viruses, and built a database named H2Flu to store and organize these genes or proteins. The database includes 28,639 differentially expressed genes (DEGs), 1,850 differentially expressed proteins, and 442 proteins with differential posttranslational modifications after influenza virus infection, as well as 3,040 human proteins that interact with influenza virus proteins and 57 human susceptibility genes. Further analysis showed that the dynamic response of human cells to virus infection, cell type and strain specificity contribute significantly to the diversity of DEGs. Additionally, large heterogeneity was also observed in protein-protein interactions between humans and different types or subtypes of influenza viruses. Overall, the study deepens our understanding of the diversity and complexity of interactions between influenza viruses and humans, and provides a valuable resource for further studies on such interactions.
Keywords: Bioinformatics; Influenza virus; Multi-omics data.
©2023 Zhu et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
Similar articles
-
Inherent Heterogeneity of Influenza A Virus Stability following Aerosolization.Appl Environ Microbiol. 2022 Feb 22;88(4):e0227121. doi: 10.1128/aem.02271-21. Epub 2022 Jan 5. Appl Environ Microbiol. 2022. PMID: 34985975 Free PMC article.
-
A comprehensive study on cellular RNA editing activity in response to infections with different subtypes of influenza a viruses.BMC Genomics. 2018 Jan 19;19(Suppl 1):925. doi: 10.1186/s12864-017-4330-1. BMC Genomics. 2018. PMID: 29363430 Free PMC article.
-
Unexpected Functional Divergence of Bat Influenza Virus NS1 Proteins.J Virol. 2018 Feb 12;92(5):e02097-17. doi: 10.1128/JVI.02097-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237829 Free PMC article.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
-
Enhancement of influenza virus transmission by gene reassortment.Curr Top Microbiol Immunol. 2014;385:185-204. doi: 10.1007/82_2014_389. Curr Top Microbiol Immunol. 2014. PMID: 25048543 Review.
Cited by
-
Raman signatures of type A and B influenza viruses: molecular origin of the "catch and kill" inactivation mechanism mediated by micrometric silicon nitride powder.RSC Chem Biol. 2025 Jan 22;6(2):182-208. doi: 10.1039/d4cb00237g. eCollection 2025 Feb 5. RSC Chem Biol. 2025. PMID: 39850321 Free PMC article.
References
-
- Canese K, Weis S. PubMed: the bibliographic database. The NCBI handbook 2(1).2013.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

