AI/ML combined with next-generation sequencing of VHH immune repertoires enables the rapid identification of de novo humanized and sequence-optimized single domain antibodies: a prospective case study
- PMID: 37842638
- PMCID: PMC10575757
- DOI: 10.3389/fmolb.2023.1249247
AI/ML combined with next-generation sequencing of VHH immune repertoires enables the rapid identification of de novo humanized and sequence-optimized single domain antibodies: a prospective case study
Abstract
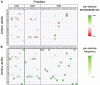
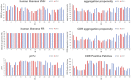
Introduction: In this study, we demonstrate the feasibility of yeast surface display (YSD) and nextgeneration sequencing (NGS) in combination with artificial intelligence and machine learning methods (AI/ML) for the identification of de novo humanized single domain antibodies (sdAbs) with favorable early developability profiles. Methods: The display library was derived from a novel approach, in which VHH-based CDR3 regions obtained from a llama (Lama glama), immunized against NKp46, were grafted onto a humanized VHH backbone library that was diversified in CDR1 and CDR2. Following NGS analysis of sequence pools from two rounds of fluorescence-activated cell sorting we focused on four sequence clusters based on NGS frequency and enrichment analysis as well as in silico developability assessment. For each cluster, long short-term memory (LSTM) based deep generative models were trained and used for the in silico sampling of new sequences. Sequences were subjected to sequence- and structure-based in silico developability assessment to select a set of less than 10 sequences per cluster for production. Results: As demonstrated by binding kinetics and early developability assessment, this procedure represents a general strategy for the rapid and efficient design of potent and automatically humanized sdAb hits from screening selections with favorable early developability profiles.
Keywords: artificial intelligence and machine learning (ML); deep learning; in silico developability; long short-term memory (LSTM); next-generation sequencing (NGS); protein engineering; single domain antibodies (VHH); yeast surface display (YSD).
Copyright © 2023 Arras, Yoo, Pekar, Clarke, Friedrich, Schröter, Schanz, Tonillo, Siegmund, Doerner, Krah, Guarnera, Zielonka and Evers.
Conflict of interest statement
PA, HY, LP, LF, VS, AD, SK, EG, SZ, AE were employed by Merck Healthcare KGaA. CS, JS, JT were employed by Merck KGaA. TC was employed by EMD Serono.
Figures





References
-
- Antibody Discovery Software (2023). Geneious biologics antibody discovery software. Available from: https://www.geneious.com/biopharma/ .
-
- Arras P., Yoo H. B., Pekar L., Schröter C., Clarke T., Krah S., et al. (2023). A library approach for the de novo high-throughput isolation of humanized VHH domains with favorable developability properties following camelid immunization. mAbs in press. 10.1080/19420862.2023.2261149 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

