Zfp335 establishes eTreg lineage and neonatal immune tolerance by targeting Hadha-mediated fatty acid oxidation
- PMID: 37843279
- PMCID: PMC10575732
- DOI: 10.1172/JCI166628
Zfp335 establishes eTreg lineage and neonatal immune tolerance by targeting Hadha-mediated fatty acid oxidation
Abstract
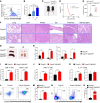
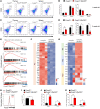
Regulatory T cells (Tregs) are instrumental in maintaining immune tolerance and preventing destructive autoimmunity, but how heterogeneous Treg populations are established remains largely unknown. Here, we show that Zfp335 deletion in Tregs failed to differentiate into effector Tregs (eTregs) and lose Treg-suppressive function and that KO mice exhibited early-onset lethal autoimmune inflammation with unrestricted activation of conventional T cells. Single-cell RNA-Seq analyses revealed that Zfp335-deficient Tregs lacked a eTreg population and showed dramatic accumulation of a dysfunctional Treg subset. Mechanistically, Zfp335-deficient Tregs displayed reduced oxidative phosphorylation and dysfunctional mitochondrial activity. Further studies revealed that Zfp335 controlled eTreg differentiation by regulating fatty acid oxidation (FAO) through direct targeting of the FAO enzyme Hadha. Importantly, we demonstrate a positive correlation between ZNF335 and HADHA expression in human eTregs. Our findings reveal that Zfp335 controls FAO-driven eTreg differentiation to establish immune tolerance.
Keywords: Adaptive immunity; Autoimmune diseases; Autoimmunity; Immunology; T cells.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

