CD8+ lymphocytes are critical for early control of tuberculosis in macaques
- PMID: 37843832
- PMCID: PMC10579699
- DOI: 10.1084/jem.20230707
CD8+ lymphocytes are critical for early control of tuberculosis in macaques
Abstract
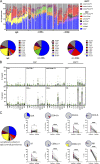
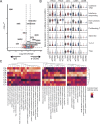
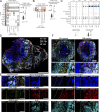
The functional role of CD8+ lymphocytes in tuberculosis remains poorly understood. We depleted innate and/or adaptive CD8+ lymphocytes in macaques and showed that loss of all CD8α+ cells (using anti-CD8α antibody) significantly impaired early control of Mycobacterium tuberculosis (Mtb) infection, leading to increased granulomas, lung inflammation, and bacterial burden. Analysis of barcoded Mtb from infected macaques demonstrated that depletion of all CD8+ lymphocytes allowed increased establishment of Mtb in lungs and dissemination within lungs and to lymph nodes, while depletion of only adaptive CD8+ T cells (with anti-CD8β antibody) worsened bacterial control in lymph nodes. Flow cytometry and single-cell RNA sequencing revealed polyfunctional cytotoxic CD8+ lymphocytes in control granulomas, while CD8-depleted animals were unexpectedly enriched in CD4 and γδ T cells adopting incomplete cytotoxic signatures. Ligand-receptor analyses identified IL-15 signaling in granulomas as a driver of cytotoxic T cells. These data support that CD8+ lymphocytes are required for early protection against Mtb and suggest polyfunctional cytotoxic responses as a vaccine target.
© 2023 Winchell et al.
Conflict of interest statement
Disclosures: J.M. Rosenberg reported personal fees from Third Rock Ventures outside the submitted work. No other disclosures were reported.
Figures










Comment in
-
Early innate role for CD8αα+ cells in tuberculosis.J Exp Med. 2023 Dec 4;220(12):e20231631. doi: 10.1084/jem.20231631. Epub 2023 Nov 2. J Exp Med. 2023. PMID: 37917028 Free PMC article.
References
-
- Balin, S.J., Pellegrini M., Klechevsky E., Won S.T., Weiss D.I., Choi A.W., Hakimian J., Lu J., Ochoa M.T., Bloom B.R., et al. . 2018. Human antimicrobial cytotoxic T lymphocytes, defined by NK receptors and antimicrobial proteins, kill intracellular bacteria. Sci. Immunol. 3:eaat7668. 10.1126/sciimmunol.aat7668 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

