Identifying Potential Lyme Disease Cases Using Self-Reported Worldwide Tweets: Deep Learning Modeling Approach Enhanced With Sentimental Words Through Emojis
- PMID: 37843893
- PMCID: PMC10616745
- DOI: 10.2196/47014
Identifying Potential Lyme Disease Cases Using Self-Reported Worldwide Tweets: Deep Learning Modeling Approach Enhanced With Sentimental Words Through Emojis
Abstract
Background: Lyme disease is among the most reported tick-borne diseases worldwide, making it a major ongoing public health concern. An effective Lyme disease case reporting system depends on timely diagnosis and reporting by health care professionals, and accurate laboratory testing and interpretation for clinical diagnosis validation. A lack of these can lead to delayed diagnosis and treatment, which can exacerbate the severity of Lyme disease symptoms. Therefore, there is a need to improve the monitoring of Lyme disease by using other data sources, such as web-based data.
Objective: We analyzed global Twitter data to understand its potential and limitations as a tool for Lyme disease surveillance. We propose a transformer-based classification system to identify potential Lyme disease cases using self-reported tweets.
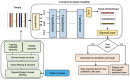
Methods: Our initial sample included 20,000 tweets collected worldwide from a database of over 1.3 million Lyme disease tweets. After preprocessing and geolocating tweets, tweets in a subset of the initial sample were manually labeled as potential Lyme disease cases or non-Lyme disease cases using carefully selected keywords. Emojis were converted to sentiment words, which were then replaced in the tweets. This labeled tweet set was used for the training, validation, and performance testing of DistilBERT (distilled version of BERT [Bidirectional Encoder Representations from Transformers]), ALBERT (A Lite BERT), and BERTweet (BERT for English Tweets) classifiers.
Results: The empirical results showed that BERTweet was the best classifier among all evaluated models (average F1-score of 89.3%, classification accuracy of 90.0%, and precision of 97.1%). However, for recall, term frequency-inverse document frequency and k-nearest neighbors performed better (93.2% and 82.6%, respectively). On using emojis to enrich the tweet embeddings, BERTweet had an increased recall (8% increase), DistilBERT had an increased F1-score of 93.8% (4% increase) and classification accuracy of 94.1% (4% increase), and ALBERT had an increased F1-score of 93.1% (5% increase) and classification accuracy of 93.9% (5% increase). The general awareness of Lyme disease was high in the United States, the United Kingdom, Australia, and Canada, with self-reported potential cases of Lyme disease from these countries accounting for around 50% (9939/20,000) of the collected English-language tweets, whereas Lyme disease-related tweets were rare in countries from Africa and Asia. The most reported Lyme disease-related symptoms in the data were rash, fatigue, fever, and arthritis, while symptoms, such as lymphadenopathy, palpitations, swollen lymph nodes, neck stiffness, and arrythmia, were uncommon, in accordance with Lyme disease symptom frequency.
Conclusions: The study highlights the robustness of BERTweet and DistilBERT as classifiers for potential cases of Lyme disease from self-reported data. The results demonstrated that emojis are effective for enrichment, thereby improving the accuracy of tweet embeddings and the performance of classifiers. Specifically, emojis reflecting sadness, empathy, and encouragement can reduce false negatives.
Keywords: BERT; Bidirectional Encoder Representations from Transformers; Lyme disease; Twitter; emojis; machine learning; natural language processing.
©Elda Kokoe Elolo Laison, Mohamed Hamza Ibrahim, Srikanth Boligarla, Jiaxin Li, Raja Mahadevan, Austen Ng, Venkataraman Muthuramalingam, Wee Yi Lee, Yijun Yin, Bouchra R Nasri. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 16.10.2023.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
Similar articles
-
Leveraging machine learning approaches for predicting potential Lyme disease cases and incidence rates in the United States using Twitter.BMC Med Inform Decis Mak. 2023 Oct 16;23(1):217. doi: 10.1186/s12911-023-02315-z. BMC Med Inform Decis Mak. 2023. PMID: 37845666 Free PMC article.
-
Comparison of pretrained transformer-based models for influenza and COVID-19 detection using social media text data in Saskatchewan, Canada.Front Digit Health. 2023 Jun 28;5:1203874. doi: 10.3389/fdgth.2023.1203874. eCollection 2023. Front Digit Health. 2023. PMID: 37448834 Free PMC article.
-
Classification of Twitter Vaping Discourse Using BERTweet: Comparative Deep Learning Study.JMIR Med Inform. 2022 Jul 21;10(7):e33678. doi: 10.2196/33678. JMIR Med Inform. 2022. PMID: 35862172 Free PMC article.
-
Evaluation of a prototype machine learning tool to semi-automate data extraction for systematic literature reviews.Syst Rev. 2023 Oct 6;12(1):187. doi: 10.1186/s13643-023-02351-w. Syst Rev. 2023. PMID: 37803451 Free PMC article.
-
Sentiment Analysis in Social Media Data for Depression Detection Using Artificial Intelligence: A Review.SN Comput Sci. 2022;3(1):74. doi: 10.1007/s42979-021-00958-1. Epub 2021 Nov 19. SN Comput Sci. 2022. PMID: 34816124 Free PMC article. Review.
Cited by
-
Identifying the geographic leading edge of Lyme disease in the United States with internet searches: A spatiotemporal analysis of Google Health Trends data.PLoS One. 2024 Nov 13;19(11):e0312277. doi: 10.1371/journal.pone.0312277. eCollection 2024. PLoS One. 2024. PMID: 39535983 Free PMC article.
-
Enhancing Automatic PT Tagging for MEDLINE Citations Using Transformer-Based Models.ArXiv [Preprint]. 2025 Jun 3:arXiv:2506.03321v1. ArXiv. 2025. PMID: 40735093 Free PMC article. Preprint.
References
-
- Wachter J, Martens C, Barbian K, Rego ROM, Rosa P. Epigenomic Landscape of Lyme Disease Spirochetes Reveals Novel Motifs. mBio. 2021 Jun 29;12(3):e0128821. doi: 10.1128/mBio.01288-21. https://journals.asm.org/doi/10.1128/mBio.01288-21?url_ver=Z39.88-2003&r... - DOI - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

