TERRA R-loops connect and protect sister telomeres in mitosis
- PMID: 37843976
- PMCID: PMC10873023
- DOI: 10.1016/j.celrep.2023.113235
TERRA R-loops connect and protect sister telomeres in mitosis
Abstract
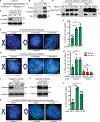
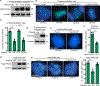
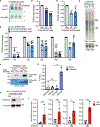
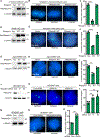
Resolution of cohesion between sister telomeres in human cells depends on TRF1-mediated recruitment of the polyADP-ribosyltransferase tankyrase to telomeres. In human aged cells, due to insufficient recruitment of TRF1/tankyrase to shortened telomeres, sisters remain cohered in mitosis. This persistent cohesion plays a protective role, but the mechanism by which sisters remain cohered is not well understood. Here we show that telomere repeat-containing RNA (TERRA) holds sister telomeres together through RNA-DNA hybrid (R-loop) structures. We show that a tankyrase-interacting partner, the RNA-binding protein C19orf43, is required for repression of TERRA R-loops. Persistent telomere cohesion in C19orf43-depleted cells is counteracted by RNaseH1, confirming that RNA-DNA hybrids hold sisters together. Consistent with a protective role for persistent telomere cohesion, depletion of C19orf43 in aged cells reduces DNA damage and delays replicative senescence. We propose that the inherent inability of shortened telomeres to recruit R-loop-repressing machinery permits a controlled onset of senescence.
Keywords: C19orf43; CP: Molecular biology; R-Loops; RNA-DNA hybrids; RNaseH; TERRA; TRF1; cohesion; senescence; tankyrase; telomeres.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests There are no competing interests.
Figures
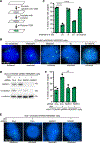
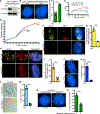
Similar articles
-
Oxidative stress at telomeres triggers internal DNA loops, TRF1 dissociation, and TRF2-dependent R-loops.Nucleic Acids Res. 2025 Apr 10;53(7):gkaf285. doi: 10.1093/nar/gkaf285. Nucleic Acids Res. 2025. PMID: 40219969 Free PMC article.
-
Persistent telomere cohesion protects aged cells from premature senescence.Nat Commun. 2020 Jul 3;11(1):3321. doi: 10.1038/s41467-020-17133-4. Nat Commun. 2020. PMID: 32620872 Free PMC article.
-
RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops.Nature. 2020 Nov;587(7833):303-308. doi: 10.1038/s41586-020-2815-6. Epub 2020 Oct 14. Nature. 2020. PMID: 33057192 Free PMC article.
-
Tankyrase function at telomeres, spindle poles, and beyond.Biochimie. 2008 Jan;90(1):83-92. doi: 10.1016/j.biochi.2007.07.012. Epub 2007 Jul 24. Biochimie. 2008. PMID: 17825467 Review.
-
Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity.Front Genet. 2015 Apr 14;6:143. doi: 10.3389/fgene.2015.00143. eCollection 2015. Front Genet. 2015. PMID: 25926849 Free PMC article. Review.
Cited by
-
Atypical R-loops in cancer: decoding molecular chaos for therapeutic gain.J Transl Med. 2025 Aug 14;23(1):912. doi: 10.1186/s12967-025-06929-x. J Transl Med. 2025. PMID: 40813661 Free PMC article. Review.
-
TERRA R-loops trigger a switch in telomere maintenance towards break-induced replication and PRIMPOL-dependent repair.EMBO J. 2025 Aug;44(16):4525-4551. doi: 10.1038/s44318-025-00502-4. Epub 2025 Jul 7. EMBO J. 2025. PMID: 40624280 Free PMC article.
-
Oxidative stress at telomeres triggers internal DNA loops, TRF1 dissociation, and TRF2-dependent R-loops.Nucleic Acids Res. 2025 Apr 10;53(7):gkaf285. doi: 10.1093/nar/gkaf285. Nucleic Acids Res. 2025. PMID: 40219969 Free PMC article.
-
RNA biogenesis and RNA metabolism factors as R-loop suppressors: a hidden role in genome integrity.Genes Dev. 2024 Jul 19;38(11-12):504-527. doi: 10.1101/gad.351853.124. Genes Dev. 2024. PMID: 38986581 Free PMC article. Review.
References
-
- de Lange T (2005). Shelterin: the protein complex that shapes and sfeguards human telomeres. Genes Dev 19, 2100–2110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

