An unconventional gatekeeper mutation sensitizes inositol hexakisphosphate kinases to an allosteric inhibitor
- PMID: 37843983
- PMCID: PMC10578927
- DOI: 10.7554/eLife.88982
An unconventional gatekeeper mutation sensitizes inositol hexakisphosphate kinases to an allosteric inhibitor
Abstract
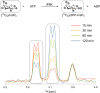
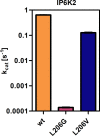
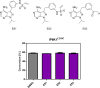
Inositol hexakisphosphate kinases (IP6Ks) are emerging as relevant pharmacological targets because a multitude of disease-related phenotypes has been associated with their function. While the development of potent IP6K inhibitors is gaining momentum, a pharmacological tool to distinguish the mammalian isozymes is still lacking. Here, we implemented an analog-sensitive approach for IP6Ks and performed a high-throughput screen to identify suitable lead compounds. The most promising hit, FMP-201300, exhibited high potency and selectivity toward the unique valine gatekeeper mutants of IP6K1 and IP6K2, compared to the respective wild-type (WT) kinases. Biochemical validation experiments revealed an allosteric mechanism of action that was corroborated by hydrogen deuterium exchange mass spectrometry measurements. The latter analysis suggested that displacement of the αC helix, caused by the gatekeeper mutation, facilitates the binding of FMP-201300 to an allosteric pocket adjacent to the ATP-binding site. FMP-201300 therefore serves as a valuable springboard for the further development of compounds that can selectively target the three mammalian IP6Ks; either as analog-sensitive kinase inhibitors or as an allosteric lead compound for the WT kinases.
Keywords: allosteric inhibitor; analog-sensitive; biochemistry; chemical biology; human; inositol phosphate; kinase.
© 2023, Aguirre et al.
Conflict of interest statement
TA, GD, SH, MN, CS, VH, AS, Jv, DF No competing interests declared
Figures






Update of
- doi: 10.1101/2023.04.26.538378
- doi: 10.7554/eLife.88982.1
- doi: 10.7554/eLife.88982.2
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

