Targeting myeloid chemotaxis to reverse prostate cancer therapy resistance
- PMID: 37844613
- PMCID: PMC10686834
- DOI: 10.1038/s41586-023-06696-z
Targeting myeloid chemotaxis to reverse prostate cancer therapy resistance
Abstract
Inflammation is a hallmark of cancer1. In patients with cancer, peripheral blood myeloid expansion, indicated by a high neutrophil-to-lymphocyte ratio, associates with shorter survival and treatment resistance across malignancies and therapeutic modalities2-5. Whether myeloid inflammation drives progression of prostate cancer in humans remain unclear. Here we show that inhibition of myeloid chemotaxis can reduce tumour-elicited myeloid inflammation and reverse therapy resistance in a subset of patients with metastatic castration-resistant prostate cancer (CRPC). We show that a higher blood neutrophil-to-lymphocyte ratio reflects tumour myeloid infiltration and tumour expression of senescence-associated mRNA species, including those that encode myeloid-chemoattracting CXCR2 ligands. To determine whether myeloid cells fuel resistance to androgen receptor signalling inhibitors, and whether inhibiting CXCR2 to block myeloid chemotaxis reverses this, we conducted an investigator-initiated, proof-of-concept clinical trial of a CXCR2 inhibitor (AZD5069) plus enzalutamide in patients with metastatic CRPC that is resistant to androgen receptor signalling inhibitors. This combination was well tolerated without dose-limiting toxicity and it decreased circulating neutrophil levels, reduced intratumour CD11b+HLA-DRloCD15+CD14- myeloid cell infiltration and imparted durable clinical benefit with biochemical and radiological responses in a subset of patients with metastatic CRPC. This study provides clinical evidence that senescence-associated myeloid inflammation can fuel metastatic CRPC progression and resistance to androgen receptor blockade. Targeting myeloid chemotaxis merits broader evaluation in other cancers.
© 2023. The Author(s).
Conflict of interest statement
A. Sharp, B.G., M.C., I.F., J.R., M.R., L.G., W.Y., S.C., K.C., A.P., C.B., G.S., J. Goodall, F.R., R. Ruddle, K.E.S., J.M., D.B., C.T., A.N., N.T., D. Westaby, J.C., M.D.F., C.Y., R.M., H.B., T.P., A.T., M.P., H.T., R. Riisnaes, A.F., P.F., S.D., A. Schlag and J.S.d.B. are employees of The ICR, which has a commercial interest in abiraterone, PARP inhibition in DNA-repair-defective cancers and PI3K–AKT pathway inhibitors. C.G. was an employee of The ICR during the conduct of this work, as well as submission, review, and revision of the manuscript, and is now an employee of Roche-Genentech. A. Sharp has received travel support from Sanofi, Roche-Genentech and Nurix, and speaker honoraria from Astellas Pharma and Merck Sharp & Dohme; has served as an advisor to DE Shaw Research and CHARM Therapeutics; and has been the chief or principal investigator of industry-sponsored clinical trials. J.S.d.B. has served on advisory boards and received fees from companies including Amgen, AstraZeneca, Astellas, Bayer, Bioxcel Therapeutics, Boehringer Ingelheim, Cellcentric, Daiichi, Eisai, Genentech/Roche, Genmab, GSK, Harpoon, Janssen, Merck Serono, Merck Sharp & Dohme, Menarini/Silicon Biosystems, Orion, Pfizer, Qiagen, Sanofi Aventis, Sierra Oncology, Taiho, Terumo and Vertex Pharmaceuticals. J.S.d.B. receives funding or other support for his research work from Daiich Sankyo, AstraZeneca, Astellas, Bayer, Cellcentric, Daiichi, Roche-Genentech, Genmab, GSK, Janssen, Merck Serono, MSD, Menarini/Silicon Biosystems, Orion, Sanofi Aventis, Sierra Oncology, Taiho, Pfizer and Vertex. J.S.d.B. was named as an inventor, with no financial interest, on US patent 8,822,438. J.S.d.B. has been the chief or principal investigator of many industry-sponsored clinical trials. S.J. has served on advisory boards and received fees from companies including Accord, AstraZeneca, Astellas, Bayer, Boston Scientific, Janssen and Pfizer; and reports research funding from Boston Scientific for other research projects. I.C. reports speaker, consultancy or advisory role activities for GSK, AstraZeneca, Novartis and MSD; travel grants from Tesaro, GSK, AstraZeneca and Janssen; and research funding (to institution as the principal investigator) from MSD, Bayer, Incyte, AstraZeneca and Vivesto. U.V. reports an advisory role (institutional) to Janssen, Astellas, Merck, MSD, Pfizer, Roche, Bayer, BMS and Novartis AAA; travel support from Janssen, Merck and Ipsen; being part of the speaker bureau for (compensated, institutional) Janssen, Astellas, Pfizer, Roche, SAMO, BMS and Ipsen; and grant funding from Fond’Action. A. Stathis serves as a principal investigator and receives institutional funding for clinical trials sponsored by AstraZeneca, Bayer, Incyte, Roche, Abbvie, ADC Therapeutics, Amgen, Cellestia, Loxo Oncology, Merck MSD, Novartis, Pfizer, Philogen and Roche; received travel grants from AstraZeneca and Incyte; served on advisory boards for Janssen and Roche; provided expert testimony to Bayer and Eli Lilly. M.D.F. received travel funding from Astellas, AstraZeneca, Pfizer, Pierre Fabre, Roche, Bristol Meiers Squibb, Novartis, MSD, Janssen and Bayer, and personal fees from Janssen, Pierre Fabre and Roche (all funding and fees were outside the submitted work). J. Gil has acted as a consultant for Unity Biotechnology, Geras Bio, Myricx Pharma and Merck KGaA. Pfizer and Unity Biotechnology have financially supported research in J. Gil’s laboratory (unrelated to the work presented here). J. Gil owns equity in Geras Bio. J. Gil is a named inventor on MRC and Imperial College patents, both related to senolytic therapies (the patents are not related to the work presented here). A.A. has been the principal investigator of industry-sponsored clinical trials sponsored by Astellas Pharma Inc., AstraZeneca, Sun Pharma Global FZE; has received consulting fees from Debiopharm; and owns shares in Oncosence. B.C., D. Waugh., R.C. and C.A. do not have any competing interest to declare.
Figures






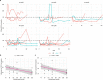


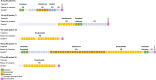

References
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

