Molecular landscape and functional characterization of centrosome amplification in ovarian cancer
- PMID: 37845213
- PMCID: PMC10579337
- DOI: 10.1038/s41467-023-41840-3
Molecular landscape and functional characterization of centrosome amplification in ovarian cancer
Abstract
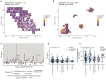
High-grade serous ovarian carcinoma (HGSOC) is characterised by poor outcome and extreme chromosome instability (CIN). Therapies targeting centrosome amplification (CA), a key mediator of chromosome missegregation, may have significant clinical utility in HGSOC. However, the prevalence of CA in HGSOC, its relationship to genomic biomarkers of CIN and its potential impact on therapeutic response have not been defined. Using high-throughput multi-regional microscopy on 287 clinical HGSOC tissues and 73 cell lines models, here we show that CA through centriole overduplication is a highly recurrent and heterogeneous feature of HGSOC and strongly associated with CIN and genome subclonality. Cell-based studies showed that high-prevalence CA is phenocopied in ovarian cancer cell lines, and that high CA is associated with increased multi-treatment resistance; most notably to paclitaxel, the commonest treatment used in HGSOC. CA in HGSOC may therefore present a potential driver of tumour evolution and a powerful biomarker for response to standard-of-care treatment.
© 2023. Springer Nature Limited.
Conflict of interest statement
J.D.B. is a co-founder and shareholder of Tailor Bio Ltd. There are no further conflicts of interests to declare.
Figures








References
-
- Morden, C. R. et al. Chromosome instability is prevalent and dynamic in high-grade serous ovarian cancer patient samples. Gynecol. Oncol.10.1016/j.ygyno.2021.02.038 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

