Genome-wide association meta-analysis identifies risk loci for abdominal aortic aneurysm and highlights PCSK9 as a therapeutic target
- PMID: 37845353
- PMCID: PMC10632148
- DOI: 10.1038/s41588-023-01510-y
Genome-wide association meta-analysis identifies risk loci for abdominal aortic aneurysm and highlights PCSK9 as a therapeutic target
Abstract
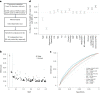
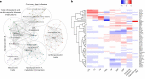
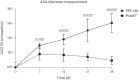
Abdominal aortic aneurysm (AAA) is a common disease with substantial heritability. In this study, we performed a genome-wide association meta-analysis from 14 discovery cohorts and uncovered 141 independent associations, including 97 previously unreported loci. A polygenic risk score derived from meta-analysis explained AAA risk beyond clinical risk factors. Genes at AAA risk loci indicate involvement of lipid metabolism, vascular development and remodeling, extracellular matrix dysregulation and inflammation as key mechanisms in AAA pathogenesis. These genes also indicate overlap between the development of AAA and other monogenic aortopathies, particularly via transforming growth factor β signaling. Motivated by the strong evidence for the role of lipid metabolism in AAA, we used Mendelian randomization to establish the central role of nonhigh-density lipoprotein cholesterol in AAA and identified the opportunity for repurposing of proprotein convertase, subtilisin/kexin-type 9 (PCSK9) inhibitors. This was supported by a study demonstrating that PCSK9 loss of function prevented the development of AAA in a preclinical mouse model.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
A.E.L. is an employee of Regeneron Genetics Center and a shareholder of Regeneron Pharmaceuticals. A.B. is an employee of Regeneron Genetics Center and a shareholder of Regeneron Pharmaceuticals. C.J.W. is currently employed by Regeneron Pharmaceuticals, but scientific input to this manuscript occurred prior to employment at Regeneron Pharmaceuticals. D.K. is a scientific advisor and reports consulting fees from Bitterroot Bio, Inc unrelated to the present work. D.L. is an employee of Regeneron Genetics Center and a shareholder of Regeneron Pharmaceuticals. D.G. is employed part-time by Novo Nordisk. J.B.N. is employed by Regeneron Pharmaceuticals, unrelated to this work. J.A.v.H. is a consultant and/or proctor for Terumo Aortic, Cook Medical, Microport, WL Gore and Philips. M.A.F. is an employee of Regeneron Genetics Center and a shareholder of Regeneron Pharmaceuticals. M.R. is a member of the Scientific Advisory Board for Cipherome. S.E.G. is currently employed by Regeneron Pharmaceuticals. S.M.D. receives research support to the University of Pennsylvania from RenalytixAI and Novo Nordisk and personal consulting fees from Calico Labs, all outside the scope of the current research. S.B. has ownerships in Intomics A/S, Hoba Therapeutics Aps, Novo Nordisk A/S, Lundbeck A/S, ALK-Abello A/S and managing board memberships in Proscion A/S and Intomics A/S. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL166991/HL/NHLBI NIH HHS/United States
- K24 HL137010/HL/NHLBI NIH HHS/United States
- R01 HL109946/HL/NHLBI NIH HHS/United States
- R01 HL059367/HL/NHLBI NIH HHS/United States
- CH/F/22/90014/BHF_/British Heart Foundation/United Kingdom
- R35 HL135824/HL/NHLBI NIH HHS/United States
- R35 HL161016/HL/NHLBI NIH HHS/United States
- MC_UU_00002/7/MRC_/Medical Research Council/United Kingdom
- T32 HL098049/HL/NHLBI NIH HHS/United States
- MR/W029790/1/MRC_/Medical Research Council/United Kingdom
- IK2 CX001780/CX/CSRD VA/United States
- IK2 BX005759/BX/BLRD VA/United States
- I01 BX003362/BX/BLRD VA/United States
- WT_/Wellcome Trust/United Kingdom
- I01 BX005831/BX/BLRD VA/United States
- MR/S019669/1/MRC_/Medical Research Council/United Kingdom
- T32 HL007843/HL/NHLBI NIH HHS/United States
- U01 HG011710/HG/NHGRI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- R01 HL142023/HL/NHLBI NIH HHS/United States
- R01 HL155209/HL/NHLBI NIH HHS/United States
- R01 HL103695/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

