Spatial transcriptomics reveals the distinct organization of mouse prefrontal cortex and neuronal subtypes regulating chronic pain
- PMID: 37845544
- PMCID: PMC10620082
- DOI: 10.1038/s41593-023-01455-9
Spatial transcriptomics reveals the distinct organization of mouse prefrontal cortex and neuronal subtypes regulating chronic pain
Erratum in
-
Author Correction: Spatial transcriptomics reveals the distinct organization of mouse prefrontal cortex and neuronal subtypes regulating chronic pain.Nat Neurosci. 2025 Oct;28(10):2167. doi: 10.1038/s41593-025-02077-z. Nat Neurosci. 2025. PMID: 40958041 Free PMC article. No abstract available.
Abstract
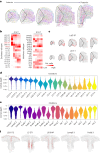
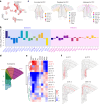
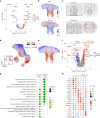
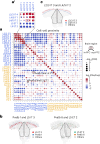
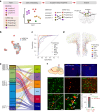
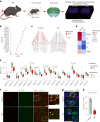
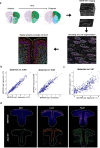
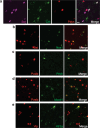
The prefrontal cortex (PFC) is a complex brain region that regulates diverse functions ranging from cognition, emotion and executive action to even pain processing. To decode the cellular and circuit organization of such diverse functions, we employed spatially resolved single-cell transcriptome profiling of the adult mouse PFC. Results revealed that PFC has distinct cell-type composition and gene-expression patterns relative to neighboring cortical areas-with neuronal excitability-regulating genes differently expressed. These cellular and molecular features are further segregated within PFC subregions, alluding to the subregion-specificity of several PFC functions. PFC projects to major subcortical targets through combinations of neuronal subtypes, which emerge in a target-intrinsic fashion. Finally, based on these features, we identified distinct cell types and circuits in PFC underlying chronic pain, an escalating healthcare challenge with limited molecular understanding. Collectively, this comprehensive map will facilitate decoding of discrete molecular, cellular and circuit mechanisms underlying specific PFC functions in health and disease.
© 2023. The Author(s).
Conflict of interest statement
J.R.M. is a cofounder of and consultant for Vizgen. The remaining authors declare no competing interests.
Figures









References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

