Leveraging machine learning approaches for predicting potential Lyme disease cases and incidence rates in the United States using Twitter
- PMID: 37845666
- PMCID: PMC10578027
- DOI: 10.1186/s12911-023-02315-z
Leveraging machine learning approaches for predicting potential Lyme disease cases and incidence rates in the United States using Twitter
Abstract
Background: Lyme disease is one of the most commonly reported infectious diseases in the United States (US), accounting for more than [Formula: see text] of all vector-borne diseases in North America.
Objective: In this paper, self-reported tweets on Twitter were analyzed in order to predict potential Lyme disease cases and accurately assess incidence rates in the US.
Methods: The study was done in three stages: (1) Approximately 1.3 million tweets were collected and pre-processed to extract the most relevant Lyme disease tweets with geolocations. A subset of tweets were semi-automatically labelled as relevant or irrelevant to Lyme disease using a set of precise keywords, and the remaining portion were manually labelled, yielding a curated labelled dataset of 77, 500 tweets. (2) This labelled data set was used to train, validate, and test various combinations of NLP word embedding methods and prominent ML classification models, such as TF-IDF and logistic regression, Word2vec and XGboost, and BERTweet, among others, to identify potential Lyme disease tweets. (3) Lastly, the presence of spatio-temporal patterns in the US over a 10-year period were studied.
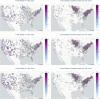
Results: Preliminary results showed that BERTweet outperformed all tested NLP classifiers for identifying Lyme disease tweets, achieving the highest classification accuracy and F1-score of [Formula: see text]. There was also a consistent pattern indicating that the West and Northeast regions of the US had a higher tweet rate over time.
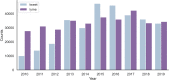
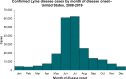
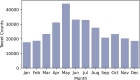
Conclusions: We focused on the less-studied problem of using Twitter data as a surveillance tool for Lyme disease in the US. Several crucial findings have emerged from the study. First, there is a fairly strong correlation between classified tweet counts and Lyme disease counts, with both following similar trends. Second, in 2015 and early 2016, the social media network like Twitter was essential in raising popular awareness of Lyme disease. Third, counties with a high incidence rate were not necessarily related with a high tweet rate, and vice versa. Fourth, BERTweet can be used as a reliable NLP classifier for detecting relevant Lyme disease tweets.
Keywords: BERT; Incidence Rate; Lyme disease; Twitter.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Murphree Bacon R, Kugeler KJ, Mead PS. Surveillance for Lyme disease--United States, 1992-2006. 2008. - PubMed
-
- Kumar D, Downs LP, Adegoke A, Machtinger E, Oggenfuss K, Ostfeld RS, et al. An Exploratory Study on the Microbiome of Northern and Southern Populations of Ixodes scapularis Ticks Predicts Changes and Unique Bacterial Interactions. Pathogens. 2022;11(2):130. 10.3390/pathogens11020130. Accessed 17 Sep 2022. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

