Single-cell RNA sequencing distinctly characterizes the wide heterogeneity in pediatric mixed phenotype acute leukemia
- PMID: 37845689
- PMCID: PMC10577904
- DOI: 10.1186/s13073-023-01241-z
Single-cell RNA sequencing distinctly characterizes the wide heterogeneity in pediatric mixed phenotype acute leukemia
Abstract
Background: Mixed phenotype acute leukemia (MPAL), a rare subgroup of leukemia characterized by blast cells with myeloid and lymphoid lineage features, is difficult to diagnose and treat. A better characterization of MPAL is essential to understand the subtype heterogeneity and how it compares with acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL). Therefore, we performed single-cell RNA sequencing (scRNAseq) on pediatric MPAL bone marrow (BM) samples to develop a granular map of the MPAL blasts and microenvironment landscape.
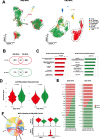
Methods: We analyzed over 40,000 cells from nine pediatric MPAL BM samples to generate a single-cell transcriptomic landscape of B/myeloid (B/My) and T/myeloid (T/My) MPAL. Cells were clustered using unsupervised single-cell methods, and malignant blast and immune clusters were annotated. Differential expression analysis was performed to identify B/My and T/My MPAL blast-specific signatures by comparing transcriptome profiles of MPAL with normal BM, AML, and ALL. Gene set enrichment analysis (GSEA) was performed, and significantly enriched pathways were compared in MPAL subtypes.
Results: B/My and T/My MPAL blasts displayed distinct blast signatures. Transcriptomic analysis revealed that B/My MPAL profile overlaps with B-ALL and AML samples. Similarly, T/My MPAL exhibited overlap with T-ALL and AML samples. Genes overexpressed in both MPAL subtypes' blast cells compared to AML, ALL, and healthy BM included MAP2K2 and CD81. Subtype-specific genes included HBEGF for B/My and PTEN for T/My. These marker sets segregated bulk RNA-seq AML, ALL, and MPAL samples based on expression profiles. Analysis comparing T/My MPAL to ETP, near-ETP, and non-ETP T-ALL, showed that T/My MPAL had greater overlap with ETP-ALL cases. Comparisons among MPAL subtypes between adult and pediatric samples showed analogous transcriptomic landscapes of corresponding subtypes. Transcriptomic differences were observed in the MPAL samples based on response to induction chemotherapy, including selective upregulation of the IL-16 pathway in relapsed samples.
Conclusions: We have for the first time described the single-cell transcriptomic landscape of pediatric MPAL and demonstrated that B/My and T/My MPAL have distinct scRNAseq profiles from each other, AML, and ALL. Differences in transcriptomic profiles were seen based on response to therapy, but larger studies will be needed to validate these findings.
Keywords: Mixed phenotype acute leukemia; Single-cell RNA sequencing; Tumor microenvironment.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
MB serves on the board of Canomiks Inc. as chief scientific advisor and has equity in it. DKG and DD hold equity in Meryx Inc. SSB serves as CEO of Anxomics LLC and has equity in it. The remaining authors declare no other competing interests.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

