Ralstonia solanacearum pandemic lineage strain UW551 overcomes inhibitory xylem chemistry to break tomato bacterial wilt resistance
- PMID: 37846613
- PMCID: PMC10782650
- DOI: 10.1111/mpp.13395
Ralstonia solanacearum pandemic lineage strain UW551 overcomes inhibitory xylem chemistry to break tomato bacterial wilt resistance
Abstract
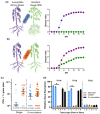
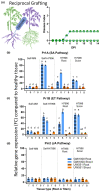
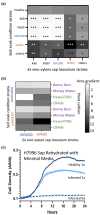
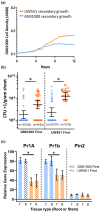
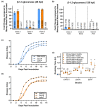
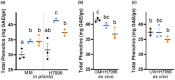
Plant-pathogenic Ralstonia strains cause bacterial wilt disease by colonizing xylem vessels of many crops, including tomato. Host resistance is the best control for bacterial wilt, but resistance mechanisms of the widely used Hawaii 7996 tomato breeding line (H7996) are unknown. Using growth in ex vivo xylem sap as a proxy for host xylem, we found that Ralstonia strain GMI1000 grows in sap from both healthy plants and Ralstonia-infected susceptible plants. However, sap from Ralstonia-infected H7996 plants inhibited Ralstonia growth, suggesting that in response to Ralstonia infection, resistant plants increase inhibitors in their xylem sap. Consistent with this, reciprocal grafting and defence gene expression experiments indicated that H7996 wilt resistance acts in both above- and belowground plant parts. Concerningly, H7996 resistance is broken by Ralstonia strain UW551 of the pandemic lineage that threatens highland tropical agriculture. Unlike other Ralstonia, UW551 grew well in sap from Ralstonia-infected H7996 plants. Moreover, other Ralstonia strains could grow in sap from H7996 plants previously infected by UW551. Thus, UW551 overcomes H7996 resistance in part by detoxifying inhibitors in xylem sap. Testing a panel of xylem sap compounds identified by metabolomics revealed that no single chemical differentially inhibits Ralstonia strains that cannot infect H7996. However, sap from Ralstonia-infected H7996 contained more phenolic compounds, which are known to be involved in plant antimicrobial defence. Culturing UW551 in this sap reduced total phenolic levels, indicating that the resistance-breaking Ralstonia strain degrades these chemical defences. Together, these results suggest that H7996 tomato wilt resistance depends in part on inducible phenolic compounds in xylem sap.
Keywords: Hawaii 7996 tomato; Ralstonia Race 3 biovar 2; Ralstonia solanacearum species complex; antimicrobial phenolics; bacterial wilt; bacterial wilt resistance.
© 2023 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures

Similar articles
-
The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato.mBio. 2012 Aug 31;3(4):e00114-12. doi: 10.1128/mBio.00114-12. Print 2012. mBio. 2012. PMID: 22807564 Free PMC article.
-
Metabolomics of tomato xylem sap during bacterial wilt reveals Ralstonia solanacearum produces abundant putrescine, a metabolite that accelerates wilt disease.Environ Microbiol. 2018 Apr;20(4):1330-1349. doi: 10.1111/1462-2920.14020. Epub 2017 Dec 22. Environ Microbiol. 2018. PMID: 29215193 Free PMC article.
-
A Single Regulator Mediates Strategic Switching between Attachment/Spread and Growth/Virulence in the Plant Pathogen Ralstonia solanacearum.mBio. 2017 Sep 26;8(5):e00895-17. doi: 10.1128/mBio.00895-17. mBio. 2017. PMID: 28951474 Free PMC article.
-
How Ralstonia solanacearum Exploits and Thrives in the Flowing Plant Xylem Environment.Trends Microbiol. 2018 Nov;26(11):929-942. doi: 10.1016/j.tim.2018.06.002. Epub 2018 Jun 22. Trends Microbiol. 2018. PMID: 29941188 Review.
-
Getting to the root of Ralstonia invasion.Semin Cell Dev Biol. 2023 Oct-Nov;148-149:3-12. doi: 10.1016/j.semcdb.2022.12.002. Epub 2022 Dec 14. Semin Cell Dev Biol. 2023. PMID: 36526528 Review.
Cited by
-
Transcriptome responses to Ralstonia solanacearum infection in tetraploid potato.Heliyon. 2025 Jan 10;11(2):e41903. doi: 10.1016/j.heliyon.2025.e41903. eCollection 2025 Jan 30. Heliyon. 2025. PMID: 39897796 Free PMC article.
-
Key mechanisms of plant-Ralstonia solanacearum interaction in bacterial wilt pathogenesis.Front Microbiol. 2025 Jun 6;16:1521422. doi: 10.3389/fmicb.2025.1521422. eCollection 2025. Front Microbiol. 2025. PMID: 40547798 Free PMC article. Review.
-
The Fusion Gene BPI-LY, Encoding Human Bactericidal/Permeability-Increasing Protein Core Fragments and Lysozyme, Enhanced the Resistance of Transgenic Tomato Plants to Bacterial Wilt.Plants (Basel). 2025 Jun 20;14(13):1897. doi: 10.3390/plants14131897. Plants (Basel). 2025. PMID: 40647907 Free PMC article.
References
-
- Ainsworth, E.A. & Gillespie, K.M. (2007) Estimation of total phenolic content and other oxidation substrates in plant tissues using Folin–Ciocalteu reagent. Nature Protocols, 2, 875–877. - PubMed
-
- Boshou, L. (2005) A broad review and perspective on breeding for resistance to bacterial wilt. In: Allen, C. , Prior, P. & Hayward, A.C. (Eds.) Bacterial wilt disease and the Ralstonia solanacearum species complex. St Paul, MN: American Phytopathology Society Press, pp. 225–238.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

