Non-β Lactam Inhibitors of the Serine β-Lactamase blaCTX-M15 in Drug-Resistant Salmonella typhi
- PMID: 37847018
- PMCID: PMC10698858
- DOI: 10.1021/acs.jcim.3c00780
Non-β Lactam Inhibitors of the Serine β-Lactamase blaCTX-M15 in Drug-Resistant Salmonella typhi
Abstract
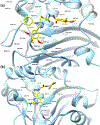
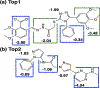
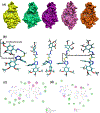
Antibiotic resistance by bacterial pathogens against widely used β-lactam drugs is a major concern to public health worldwide, resulting in high healthcare cost. The present study aimed to extend previous research by investigating the potential activity of reported compounds against the S. typhi β-lactamase protein. 74 compounds from computational screening reported in our previous study against β-lactamase CMY-10 were subjected to docking studies against blaCTX-M15. Site-Identification by Ligand Competitive Saturation (SILCS)-Monte Carlo (SILCS-MC) was applied to the top two ligands selected from molecular docking studies to predict and refine their conformations for binding conformations against blaCTX-M15. The SILCS-MC method predicted affinities of -8.6 and -10.7 kcal/mol for Top1 and Top2, respectively, indicating low micromolar binding to the blaCTX-M15 active site. MD simulations initiated from SILCS-MC docked orientations were carried out to better characterize the dynamics and stability of the complexes. Important interactions anchoring the ligand within the active site include pi-pi stacked, amide-pi, and pi-alkyl interactions. Simulations of the Top2-blaCTX-M15 complex exhibited stability associated with a wide range of hydrogen-bond and aromatic interactions between the protein and the ligand. Experimental β-lactamase (BL) activity assays showed that Top1 has 0.1 u/mg BL activity, and Top2 has a BL activity of 0.038 u/mg with a minimum inhibitory concentration of 1 mg/mL. The inhibitors proposed in this study are non-β-lactam-based β-lactamase inhibitors that exhibit the potential to be used in combination with β-lactam antibiotics against multidrug-resistant clinical isolates. Thus, Top1 and Top2 represent lead compounds that increase the efficacy of β-lactam antibiotics with a low dose concentration.
Conflict of interest statement
Conflict of Interest
ADM is co-founder and CSO of SilcsBio LLC.
Figures





References
-
- Wong VK; Baker S; Pickard DJ; Parkhill J; Page AJ; Feasey NA; Kingsley RA; Thomson NR; Keane JA; Weill F-X; Edwards DJ; Hawkey J; Harris SR; Mather AE; Cain AK; Hadfield J; Hart PJ; Thieu NTV; Klemm EJ; Glinos DA; Breiman RF; Watson CH; Kariuki S; Gordon MA; Heyderman RS; Okoro C; Jacobs J; Lunguya O; Edmunds WJ; Msefula C; Chabalgoity JA; Kama M; Jenkins K; Dutta S; Marks F; Campos J; Thompson C; Obaro S; MacLennan CA; Dolecek C; Keddy KH; Smith AM; Parry CM; Karkey A; Mulholland EK; Campbell JI; Dongol S; Basnyat B; Dufour M; Bandaranayake D; Naseri TT; Singh SP; Hatta M; Newton P; Onsare RS; Isaia L; Dance D; Davong V; Thwaites G; Wijedoru L; Crump JA; De Pinna E; Nair S; Nilles EJ; Thanh DP; Turner P; Soeng S; Valcanis M; Powling J; Dimovski K; Hogg G; Farrar J; Holt KE; Dougan G Phylogeographical Analysis of the Dominant Multidrug-Resistant H58 Clade of Salmonella Typhi Identifies Inter- and Intracontinental Transmission Events. Nat. Genet 2015, 47, 632–639. - PMC - PubMed
-
- Feasey NA; Gaskell K; Wong V; Msefula C; Selemani G; Kumwenda S; Allain TJ; Mallewa J; Kennedy N; Bennett A; Nyirongo JO; Nyondo PA; Zulu MD; Parkhill J; Dougan G; Gordon MA; Heyderman RS Rapid Emergence of Multidrug Resistant, H58-Lineage Salmonella Typhi in Blantyre, Malawi. PLoS Negl. Trop. Dis 2015, 9, e0003748. - PMC - PubMed
-
- Pham Thanh D; Karkey A; Dongol S; Ho Thi N; Thompson CN; Rabaa MA; Arjyal A; Holt KE; Wong V; Tran Vu Thieu N; Voong Vinh P; Ha Thanh T; Pradhan A; Shrestha SK; Gajurel D; Pickard D; Parry CM; Dougan G; Wolbers M; Dolecek C; Thwaites GE; Basnyat B; Baker S A Novel Ciprofloxacin-Resistant Subclade of H58 Salmonella Typhi Is Associated with Fluoroquinolone Treatment Failure. eLife 2016, 5, e14003. - PMC - PubMed
-
- Chatham-Stephens K; Medalla F; Hughes M; Appiah GD; Aubert RD; Caidi H; Angelo KM; Walker AT; Hatley N; Masani S; Nash J; Belko J; Ryan ET; Mintz E; Friedman CR Emergence of Extensively Drug-Resistant Salmonella Typhi Infections Among Travelers to or from Pakistan - United States, 2016–2018. MMWR Morb Mortal Wkly Rep 2019, 68, 11–13. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

