Mathematical solutions in internal dose assessment: A comparison of Python-based differential equation solvers in biokinetic modeling
- PMID: 37848023
- PMCID: PMC10613827
- DOI: 10.1088/1361-6498/ad0409
Mathematical solutions in internal dose assessment: A comparison of Python-based differential equation solvers in biokinetic modeling
Abstract
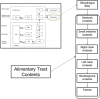
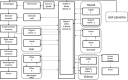
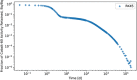
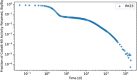
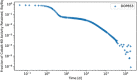
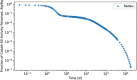
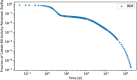
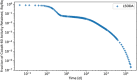
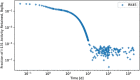
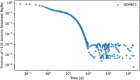
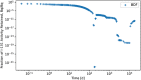
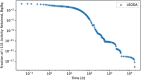
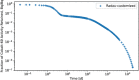
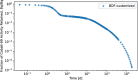
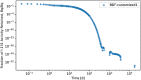
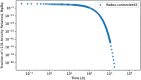
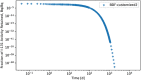
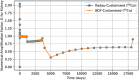
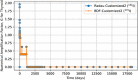
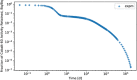
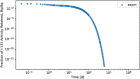
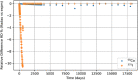
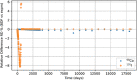
In biokinetic modeling systems employed for radiation protection, biological retention and excretion have been modeled as a series of discretized compartments representing the organs and tissues of the human body. Fractional retention and excretion in these organ and tissue systems have been mathematically governed by a series of coupled first-order ordinary differential equations (ODEs). The coupled ODE systems comprising the biokinetic models are usually stiff due to the severe difference between rapid and slow transfers between compartments. In this study, the capabilities of solving a complex coupled system of ODEs for biokinetic modeling were evaluated by comparing different Python programming language solvers and solving methods with the motivation of establishing a framework that enables multi-level analysis. The stability of the solvers was analyzed to select the best performers for solving the biokinetic problems. A Python-based linear algebraic method was also explored to examine how the numerical methods deviated from an analytical or semi-analytical method. Results demonstrated that customized implicit methods resulted in an enhanced stable solution for the inhaled60Co (Type M) and131I (Type F) exposure scenarios for the inhalation pathway of the International Commission on Radiological Protection (ICRP) Publication 130 Human Respiratory Tract Model (HRTM). The customized implementation of the Python-based implicit solvers resulted in approximately consistent solutions with the Python-based matrix exponential method (expm). The differences generally observed between the implicit solvers andexpmare attributable to numerical precision and the order of numerical approximation of the numerical solvers. This study provides the first analysis of a list of Python ODE solvers and methods by comparing their usage for solving biokinetic models using the ICRP Publication 130 HRTM and provides a framework for the selection of the most appropriate ODE solvers and methods in Python language to implement for modeling the distribution of internal radioactivity.
Keywords: ODE solvers; Python; biokinetic modeling; compartmental analysis; internal dosimetry.
Creative Commons Attribution license.
Figures




References
-
- Al-Mohy A H, Higham N J. A new scaling and squaring algorithm for the matrix exponential. SIAM J. Matrix Anal. Appl. 2010;31:970–89. doi: 10.1137/09074721X. - DOI
-
- Anderson D H.1983Compartmental modeling and tracer kinetics Lecture notes in Biomathematics edW Bossert.et alSpringer
-
- Arioli M, Codenotti B, Fassino C. The Padé method for computing the matrix exponential. Linear Algebra Appl. 1996;240:111–30. doi: 10.1016/0024-3795(94)00190-1. - DOI
-
- Aro C J. CHEMSODE: a stiff ODE solver for the equations of chemical kinetics. Comput. Phys. Commun. 1996;97:304–14. doi: 10.1016/0010-4655(96)00071-9. - DOI
-
- Ball S J, Adams R K. ‘MATEXP’, A general purpose digital computer program for solving ordinary differential equations by the matrix exponential method. 1967 (Oak Ridge National Laboratory)
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
