Rapid and sensitive one-tube detection of mpox virus using RPA-coupled CRISPR-Cas12 assay
- PMID: 37848032
- PMCID: PMC10626268
- DOI: 10.1016/j.crmeth.2023.100620
Rapid and sensitive one-tube detection of mpox virus using RPA-coupled CRISPR-Cas12 assay
Abstract
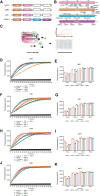
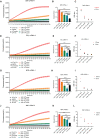
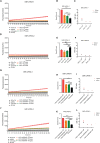
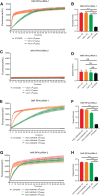
Mpox is caused by a zoonotic virus belonging to the Orthopoxvirus genus and the Poxviridae family. In this study, we develop a recombinase polymerase amplification (RPA)-coupled CRISPR-Cas12a detection assay for the mpox virus. We design and test a series of CRISPR-derived RNAs(crRNAs) targeting the conserved D6R and E9L genes for orthopoxvirus and the unique N3R and N4R genes for mpox viruses. D6R crRNA-1 exhibits the most robust activity in detecting orthopoxviruses, and N4R crRNA-2 is able to distinguish the mpox virus from other orthopoxviruses. The Cas12a/crRNA assay alone presents a detection limit of 108 copies of viral DNA, whereas coupling RPA increases the detection limit to 1-10 copies. The one-tube RPA-Cas12a assay can, therefore, detect viral DNA as low as 1 copy within 30 min and holds the promise of providing point-of-care detection for mpox viral infection.
Keywords: AsCas12a; CP: Biotechnology; CP: Microbiology; CRISPR; RPA; mpox detection; one-tube.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests F.G., F.Z., Y. Hu, F.X., and S.M. are inventors of pending and issued patents on CRISPR-Cas12a detection of mpox.
Figures

Similar articles
-
CRISPR-based strategies for sample-to-answer monkeypox detection: current status and emerging opportunities.Nanotechnology. 2024 Nov 4;36(4):042001. doi: 10.1088/1361-6528/ad892b. Nanotechnology. 2024. PMID: 39433062 Free PMC article. Review.
-
Development of a CRISPR/Cas12a-recombinase polymerase amplification assay for visual and highly specific identification of the Congo Basin and West African strains of mpox virus.J Med Virol. 2023 May;95(5):e28757. doi: 10.1002/jmv.28757. J Med Virol. 2023. PMID: 37212293
-
Amplification-free detection of Mpox virus DNA using Cas12a and multiple crRNAs.Mikrochim Acta. 2024 Jan 17;191(2):102. doi: 10.1007/s00604-024-06184-9. Mikrochim Acta. 2024. PMID: 38231433
-
CRISPR-Cas12a assisted specific detection of mpox virus.J Med Virol. 2023 Aug;95(8):e28974. doi: 10.1002/jmv.28974. J Med Virol. 2023. PMID: 37515526
-
The current status and future prospects of CRISPR-based detection of monkeypox virus: A review.Anal Chim Acta. 2025 Jan 22;1336:343295. doi: 10.1016/j.aca.2024.343295. Epub 2024 Oct 1. Anal Chim Acta. 2025. PMID: 39788645 Review.
Cited by
-
CRISPR-based strategies for sample-to-answer monkeypox detection: current status and emerging opportunities.Nanotechnology. 2024 Nov 4;36(4):042001. doi: 10.1088/1361-6528/ad892b. Nanotechnology. 2024. PMID: 39433062 Free PMC article. Review.
-
Virology, epidemiology, transmissions, diagnostic tests, prophylaxis and treatments of human Mpox: Saudi Arabia perspective.Front Cell Infect Microbiol. 2025 Feb 28;15:1530900. doi: 10.3389/fcimb.2025.1530900. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40093536 Free PMC article. Review.
-
Detection of the mpox virus using a robust recombinase-aided amplification-based approach.Biosaf Health. 2025 Mar 22;7(2):103-109. doi: 10.1016/j.bsheal.2025.03.001. eCollection 2025 Apr. Biosaf Health. 2025. PMID: 40453475 Free PMC article.
-
An ultrasensitive and specific CRISPR-Cas13a-mediated point-of-care assay for monkeypox detection and PCR-based clade detection.Infect Dis Poverty. 2025 Jun 23;14(1):56. doi: 10.1186/s40249-025-01325-5. Infect Dis Poverty. 2025. PMID: 40551276 Free PMC article.
-
Mpox disease, diagnosis, and point of care platforms.Bioeng Transl Med. 2025 Jan 2;10(3):e10733. doi: 10.1002/btm2.10733. eCollection 2025 May. Bioeng Transl Med. 2025. PMID: 40385539 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

