Streamlined structure determination by cryo-electron tomography and subtomogram averaging using TomoBEAR
- PMID: 37848413
- PMCID: PMC10582028
- DOI: 10.1038/s41467-023-42085-w
Streamlined structure determination by cryo-electron tomography and subtomogram averaging using TomoBEAR
Erratum in
-
Author Correction: Streamlined structure determination by cryo-electron tomography and subtomogram averaging using TomoBEAR.Nat Commun. 2024 Jul 3;15(1):5594. doi: 10.1038/s41467-024-49476-7. Nat Commun. 2024. PMID: 38961102 Free PMC article. No abstract available.
Abstract
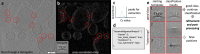
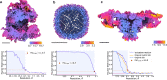
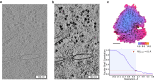
Structures of macromolecules in their native state provide unique unambiguous insights into their functions. Cryo-electron tomography combined with subtomogram averaging demonstrated the power to solve such structures in situ at resolutions in the range of 3 Angstrom for some macromolecules. In order to be applicable to the structural determination of the majority of macromolecules observable in cells in limited amounts, processing of tomographic data has to be performed in a high-throughput manner. Here we present TomoBEAR-a modular configurable workflow engine for streamlined processing of cryo-electron tomographic data for subtomogram averaging. TomoBEAR combines commonly used cryo-EM packages with reasonable presets to provide a transparent ("white box") approach for data management and processing. We demonstrate applications of TomoBEAR to two data sets of purified macromolecular targets, to an ion channel RyR1 in a membrane, and the tomograms of plasma FIB-milled lamellae and demonstrate the ability to produce high-resolution structures. TomoBEAR speeds up data processing, minimizes human interventions, and will help accelerate the adoption of in situ structural biology by cryo-ET. The source code and the documentation are freely available.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Beck M, Baumeister W. Cryo-electron tomography: can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol. 2016;26:825–837. - PubMed
-
- Wan W, Briggs JAG. Cryo-electron tomography and subtomogram averaging. Methods Enzymol. 2016;579:329–367. - PubMed
-
- Leigh KE, et al. Subtomogram averaging from cryo-electron tomograms. Methods Cell Biol. 2019;152:217–259. - PubMed
-
- Schur FKM, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506–508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

