CRISPR/Cas9 mediated editing of pheromone biosynthesis activating neuropeptide (PBAN) gene disrupts mating in the Fall armyworm, Spodoptera frugiperda (J. E. Smith) (Lepidoptera: Noctuidae)
- PMID: 37849767
- PMCID: PMC10577122
- DOI: 10.1007/s13205-023-03798-3
CRISPR/Cas9 mediated editing of pheromone biosynthesis activating neuropeptide (PBAN) gene disrupts mating in the Fall armyworm, Spodoptera frugiperda (J. E. Smith) (Lepidoptera: Noctuidae)
Abstract
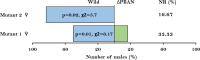
The Fall armyworm, Spodoptera frugiperda, is a globally important invasive pest, primarily on corn, causing severe yield loss. Overuse of synthetic chemicals has caused significant ecological harm, and in many instances control has failed. Therefore, developing efficient, environmentally friendly substitutes for sustainable management of this pest is of high priority. CRISPR/Cas9-mediated gene editing causes site-specific mutations that typically result in loss-of-function of the target gene. In this regard, identifying key genes that govern the reproduction of S. frugiperda and finding ways to introduce mutations in the key genes is very important for successfully managing this pest. In this study, the pheromone biosynthesis activator neuropeptide (PBAN) gene of S. frugiperda was cloned and tested for its function via a loss-of-function approach using CRISPR/Cas9. Ribonucleoprotein (RNP) complex (single guide RNA (sgRNA) targeting the PBAN gene + Cas9 protein) was validated through in vitro restriction assay followed by embryonic microinjection into the G0 stage for in vivo editing of the target gene. Specific suppression of PBAN by CRISPR/Cas9 in females significantly affected mating. Mating studies between wild males and mutant females resulted in no fecundity. This was in contrast to when mutant males were crossed with wild females, which resulted in reduced fecundity. These results suggest that mating disruption is more robust where PBAN is edited in females. The behavioural bioassay using an olfactometer revealed that mutant females were less attractive to wild males compared to wild females. This study is the first of its kind, supporting CRISPR/Cas9 mediating editing of the PBAN gene disrupting mating in S. frugiperda. Understanding the potential use of these molecular techniques may help develop novel management strategies that target other key functional genes.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-023-03798-3.
Keywords: CRISPR/Cas9; Genome editing; Mating disruption; PBAN; Spodoptera frugiperda.
© King Abdulaziz City for Science and Technology 2023. Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest in the publication.
Figures







References
-
- Altstein M, Gazit Y, Aziz OB, Gabay T, Marcus R, Vogel Z, Barg J. Induction of cuticular melanization in Spodoptera littoralis larvae by PBAN/MRCH: development of a quantitative bioassay and structure function analysis. Arch Insect Biochem. 1996;31:355–370. doi: 10.1002/(SICI)1520-6327(1996)31:4<355::AID-ARCH1>3.0.CO;2-V. - DOI
-
- Ashok K, Balasubramani V, Kennedy JS, Geethalakshmi V, Jeyakumar P, Sathiah N. Evaluating artificial diets for the fall armyworm, Spodoptera frugiperda (JE Smith) (Lepidoptera: Noctuidae) through nutritional indices and an age-stage, two-sex life table approach. Afr Entomol. 2021;29:620–634. doi: 10.4001/003.029.0620. - DOI
-
- Ashok K, Bhargava CN, Babu KP, Rohan W, Manamohan M, Rai A, Sanjay KP, Parvathy MS, Kennedy JS, Asokan R. First report on CRISPR/Cas9 mediated editing of the eye colour gene, Tryptophan 2, 3-dioxygenase in egg plant shoot and fruit borer Leucinodes orbonalis Guenée (Lepidoptera: Crambidae) J Asia-Pac Entomol. 2023;26:102031. doi: 10.1016/j.aspen.2022.102031. - DOI
-
- Bajracharya ASR, Bhat B, Sharma P, Shashank PR, Meshram NM, Hashmi TR. First record of fall army worm Spodoptera frugiperda (JE Smith) from Nepal. Indian J Entomol. 2019;81:635–639. doi: 10.5958/0974-8172.2019.00137.8. - DOI
LinkOut - more resources
Full Text Sources

