MethMotif.Org 2024: a database integrating context-specific transcription factor-binding motifs with DNA methylation patterns
- PMID: 37850642
- PMCID: PMC10767921
- DOI: 10.1093/nar/gkad894
MethMotif.Org 2024: a database integrating context-specific transcription factor-binding motifs with DNA methylation patterns
Abstract
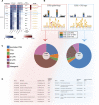
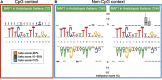
MethMotif (https://methmotif.org) is a publicly available database that provides a comprehensive repository of transcription factor (TF)-binding profiles, enriched with DNA methylation patterns. In this release, we have enhanced the platform, expanding our initial collection to over 700 position weight matrices (PWM), all of which include DNA methylation profiles. One of the key advancements in this release is the segregation of TF-binding motifs based on their cofactors and DNA methylation status. We have previously demonstrated that gene ontology (GO) enriched terms associated with TF target genes may differ based on their association with alternative cofactors and DNA methylation status. MethMotif provides precomputed GO annotations for each human TF of interest, as well as for TF-co-TF complexes, enabling a comprehensive analysis of TF functions in the context of their co-factors. Additionally, MethMotif has been updated to encompass data for two new species, Mus musculus and Arabidopsis thaliana, widening its applicability to a broader community. MethMotif stands out as the first and only TF-binding motifs database to incorporate context-specific PWM coupled with epigenetic information, thereby enlightening context-specific TF functions. This enhancement allows the community to explore and gain deeper insights into the regulatory mechanisms governing transcriptional processes.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

