metaGOflow: a workflow for the analysis of marine Genomic Observatories shotgun metagenomics data
- PMID: 37850871
- PMCID: PMC10583283
- DOI: 10.1093/gigascience/giad078
metaGOflow: a workflow for the analysis of marine Genomic Observatories shotgun metagenomics data
Abstract
Background: Genomic Observatories (GOs) are sites of long-term scientific study that undertake regular assessments of the genomic biodiversity. The European Marine Omics Biodiversity Observation Network (EMO BON) is a network of GOs that conduct regular biological community samplings to generate environmental and metagenomic data of microbial communities from designated marine stations around Europe. The development of an effective workflow is essential for the analysis of the EMO BON metagenomic data in a timely and reproducible manner.
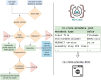
Findings: Based on the established MGnify resource, we developed metaGOflow. metaGOflow supports the fast inference of taxonomic profiles from GO-derived data based on ribosomal RNA genes and their functional annotation using the raw reads. Thanks to the Research Object Crate packaging, relevant metadata about the sample under study, and the details of the bioinformatics analysis it has been subjected to, are inherited to the data product while its modular implementation allows running the workflow partially. The analysis of 2 EMO BON samples and 1 Tara Oceans sample was performed as a use case.
Conclusions: metaGOflow is an efficient and robust workflow that scales to the needs of projects producing big metagenomic data such as EMO BON. It highlights how containerization technologies along with modern workflow languages and metadata package approaches can support the needs of researchers when dealing with ever-increasing volumes of biological data. Despite being initially oriented to address the needs of EMO BON, metaGOflow is a flexible and easy-to-use workflow that can be broadly used for one-sample-at-a-time analysis of shotgun metagenomics data.
Keywords: Common Workflow Language (CWL); MGnify; RO-Crate; containers; provenance; shotgun metagenomics.
© The Author(s) 2023. Published by Oxford University Press GigaScience.
Conflict of interest statement
M.B., L.R., and R.D.F. are members of the MGnify group that is part of the ELIXIR infrastructure [95]. The authors declare that they have no other competing interests.
Figures


References
-
- Caruso G, Azzaro M, Caroppo C, et al. Microbial community and its potential as descriptor of environmental status. ICES J Mar Sci. 2016;73(9):2174–7.. 10.1093/icesjms/fsw101. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

