A chromosome-level reference genome for the common octopus, Octopus vulgaris (Cuvier, 1797)
- PMID: 37850903
- PMCID: PMC10700109
- DOI: 10.1093/g3journal/jkad220
A chromosome-level reference genome for the common octopus, Octopus vulgaris (Cuvier, 1797)
Abstract
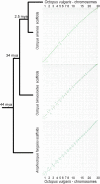
Cephalopods are emerging animal models and include iconic species for studying the link between genomic innovations and physiological and behavioral complexities. Coleoid cephalopods possess the largest nervous system among invertebrates, both for cell counts and brain-to-body ratio. Octopus vulgaris has been at the center of a long-standing tradition of research into diverse aspects of cephalopod biology, including behavioral and neural plasticity, learning and memory recall, regeneration, and sophisticated cognition. However, no chromosome-scale genome assembly was available for O. vulgaris to aid in functional studies. To fill this gap, we sequenced and assembled a chromosome-scale genome of the common octopus, O. vulgaris. The final assembly spans 2.8 billion basepairs, 99.34% of which are in 30 chromosome-scale scaffolds. Hi-C heatmaps support a karyotype of 1n = 30 chromosomes. Comparisons with other octopus species' genomes show a conserved octopus karyotype and a pattern of local genome rearrangements between species. This new chromosome-scale genome of O. vulgaris will further facilitate research in all aspects of cephalopod biology, including various forms of plasticity and the neural machinery underlying sophisticated cognition, as well as an understanding of cephalopod evolution.
Keywords: Hi-C; Octopus vulgaris; chromosome-scale; coleoid cephalopods.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest D.T.S. is a shareholder of Pacific Biosciences of California, Inc. All other authors declare no competing interests.
Figures

Similar articles
-
The survey and reference assisted assembly of the Octopus vulgaris genome.Sci Data. 2019 Apr 1;6(1):13. doi: 10.1038/s41597-019-0017-6. Sci Data. 2019. PMID: 30931949 Free PMC article.
-
The octopus genome and the evolution of cephalopod neural and morphological novelties.Nature. 2015 Aug 13;524(7564):220-4. doi: 10.1038/nature14668. Nature. 2015. PMID: 26268193 Free PMC article.
-
The gold-ringed octopus (Amphioctopus fangsiao) genome and cerebral single-nucleus transcriptomes provide insights into the evolution of karyotype and neural novelties.BMC Biol. 2022 Dec 27;20(1):289. doi: 10.1186/s12915-022-01500-2. BMC Biol. 2022. PMID: 36575497 Free PMC article.
-
Octopus vulgaris: An Alternative in Evolution.Results Probl Cell Differ. 2018;65:585-598. doi: 10.1007/978-3-319-92486-1_26. Results Probl Cell Differ. 2018. PMID: 30083937 Review.
-
Cadherin genes and evolutionary novelties in the octopus.Semin Cell Dev Biol. 2017 Sep;69:151-157. doi: 10.1016/j.semcdb.2017.06.007. Epub 2017 Jun 13. Semin Cell Dev Biol. 2017. PMID: 28627384 Review.
Cited by
-
Decoding Octopus Skin Mucus: Impact of Aquarium-Maintenance and Senescence on the Proteome Profile of the Common Octopus (Octopus vulgaris).Int J Mol Sci. 2024 Sep 15;25(18):9953. doi: 10.3390/ijms25189953. Int J Mol Sci. 2024. PMID: 39337441 Free PMC article.
-
The neural basis of visual processing and behavior in cephalopods.Curr Biol. 2023 Oct 23;33(20):R1106-R1118. doi: 10.1016/j.cub.2023.08.093. Curr Biol. 2023. PMID: 37875093 Free PMC article. Review.
-
A chromosome-level genome assembly of the Bullacta exarata (Cephalaspidea: Haminoeidae).Sci Data. 2025 Jun 16;12(1):1008. doi: 10.1038/s41597-025-05373-2. Sci Data. 2025. PMID: 40523891 Free PMC article.
-
Population genomics of a sailing siphonophore reveals genetic structure in the open ocean.Curr Biol. 2025 Aug 4;35(15):3556-3569.e6. doi: 10.1016/j.cub.2025.05.066. Epub 2025 Jun 19. Curr Biol. 2025. PMID: 40541187
-
Octopus vulgaris Exhibits Interindividual Differences in Behavioural and Problem-Solving Performance.Biology (Basel). 2023 Dec 4;12(12):1487. doi: 10.3390/biology12121487. Biology (Basel). 2023. PMID: 38132313 Free PMC article.
References
-
- Adachi K, Ohnishi K, Kuramochi T, Yoshinaga T, Okumura S-I. 2014. Molecular cytogenetic study in Octopus (Amphioctopus) areolatus from Japan. Fish Sci. 80(3):445–450. doi:10.1007/s12562-014-0703-4. - DOI
-
- Aho AV, Kernighan BW, Weinberger PJ. 1988. The AWK Programming Language. Reading (MA): Addison-Wesley.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
