Continuing genomic evolution of the Neisseria meningitidis cc11.2 urethritis clade, Nm UC: a narrative review
- PMID: 37850987
- PMCID: PMC10634446
- DOI: 10.1099/mgen.0.001113
Continuing genomic evolution of the Neisseria meningitidis cc11.2 urethritis clade, Nm UC: a narrative review
Abstract
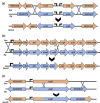
Neisseria meningitidis (Nm) is a bacterial pathogen responsible for invasive meningococcal disease. Though typically colonizing the nasopharynx, multiple outbreaks of meningococcal urethritis were first reported in 2015-2016; outbreaks originally presumed to be caused by Neisseria gonorrhoeae (Ng). Genomic analysis revealed that the Nm isolates causing these outbreaks were a distinct clade, and had integrated gonococcal DNA at multiple genomic sites, including the gonococcal denitrification apparatus aniA-norB, a partial gonococcal operon of five genes containing ispD, and the acetylglutamate kinase gene argB with the adjacent gonococcal locus NGO0843. The urethritis isolates had also deleted the group C capsule biosynthesis genes cssA/B/C and csc, resulting in loss of capsule. Collectively, these isolates form the N. meningitidis urethritis clade (NmUC). Genomic analysis of recent (2016-2022) NmUC isolates revealed that the genomic features have been maintained in the clade, implying that they are important for NmUC's status as a urogenital pathogen. Furthermore, the analysis revealed the emergence of a sub-clade, designated NmUC-B, phylogenetically separated from the earlier NmUC-A. This sub-clade has integrated additional gonococcal alleles into the genome, including alleles associated with antimicrobial resistance. NmUC continues to adapt to a urethral niche and evolve as a urogenital pathogen.
Keywords: Neisseria gonorrhoeae; Neisseria meningitidis; NmUC; antimicrobial resistance; genomic evolution; urogenital pathogen.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
-
- Cohn AC, MacNeil JR, Clark TA, Ortega-Sanchez IR, Briere EZ, et al. Prevention and control of meningococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2013;62:1–28. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

