A PINK1 input threshold arises from positive feedback in the PINK1/Parkin mitophagy decision circuit
- PMID: 37851575
- PMCID: PMC10668033
- DOI: 10.1016/j.celrep.2023.113260
A PINK1 input threshold arises from positive feedback in the PINK1/Parkin mitophagy decision circuit
Abstract
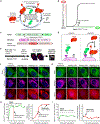
Mechanisms that prevent accidental activation of the PINK1/Parkin mitophagy circuit on healthy mitochondria are poorly understood. On the surface of damaged mitochondria, PINK1 accumulates and acts as the input signal to a positive feedback loop of Parkin recruitment, which in turn promotes mitochondrial degradation via mitophagy. However, PINK1 is also present on healthy mitochondria, where it could errantly recruit Parkin and thereby activate this positive feedback loop. Here, we explore emergent properties of the PINK1/Parkin circuit by quantifying the relationship between mitochondrial PINK1 concentrations and Parkin recruitment dynamics. We find that Parkin is recruited to mitochondria only if PINK1 levels exceed a threshold and then only after a delay that is inversely proportional to PINK1 levels. Furthermore, these two regulatory properties arise from the input-coupled positive feedback topology of the PINK1/Parkin circuit. These results outline an intrinsic mechanism by which the PINK1/Parkin circuit can avoid errant activation on healthy mitochondria.
Keywords: CP: Molecular biology; PINK1; Parkin; circuit; delay; mathematical model; mitophagy decision; quantitative microscopy; synthetic biology; systems biology; threshold.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.J.A. and L.F.W. are founders and scientific advisory board members of Nine Square Therapeutics.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

