CD73 controls Myosin II-driven invasion, metastasis, and immunosuppression in amoeboid pancreatic cancer cells
- PMID: 37851808
- PMCID: PMC10584351
- DOI: 10.1126/sciadv.adi0244
CD73 controls Myosin II-driven invasion, metastasis, and immunosuppression in amoeboid pancreatic cancer cells
Abstract
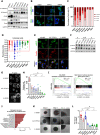
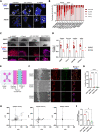
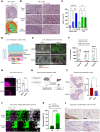
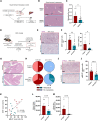
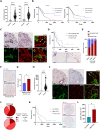
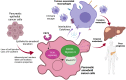
Pancreatic ductal adenocarcinoma (PDAC) has a very poor prognosis because of its high propensity to metastasize and its immunosuppressive microenvironment. Using a panel of pancreatic cancer cell lines, three-dimensional (3D) invasion systems, microarray gene signatures, microfluidic devices, mouse models, and intravital imaging, we demonstrate that ROCK-Myosin II activity in PDAC cells supports a transcriptional program conferring amoeboid invasive and immunosuppressive traits and in vivo metastatic abilities. Moreover, we find that immune checkpoint CD73 is highly expressed in amoeboid PDAC cells and drives their invasive, metastatic, and immunomodulatory traits. Mechanistically, CD73 activates RhoA-ROCK-Myosin II downstream of PI3K. Tissue microarrays of human PDAC biopsies combined with bioinformatic analysis reveal that rounded-amoeboid invasive cells with high CD73-ROCK-Myosin II activity and their immunosuppressive microenvironment confer poor prognosis to patients. We propose targeting amoeboid PDAC cells as a therapeutic strategy.
Figures


References
-
- L. Rahib, B. D. Smith, R. Aizenberg, A. B. Rosenzweig, J. M. Fleshman, L. M. Matrisian, Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 74, 2913–2921 (2014). - PubMed
-
- J. Kleeff, M. Korc, M. Apte, C. La Vecchia, C. D. Johnson, A. V. Biankin, R. E. Neale, M. Tempero, D. A. Tuveson, R. H. Hruban, J. P. Neoptolemos, Pancreatic cancer. Nat. Rev. Dis. Primer 2, 16022 (2016). - PubMed
-
- C. L. Chaffer, R. A. Weinberg, A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011). - PubMed
-
- P. Friedl, S. Alexander, Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell 147, 992–1009 (2011). - PubMed
-
- A. G. Clark, D. M. Vignjevic, Modes of cancer cell invasion and the role of the microenvironment. Curr. Opin. Cell Biol. 36, 13–22 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

