High-resolution melting analysis for simultaneous detection and discrimination between wild-type and vaccine strains of feline calicivirus
- PMID: 37851857
- PMCID: PMC11003490
- DOI: 10.1080/01652176.2023.2272188
High-resolution melting analysis for simultaneous detection and discrimination between wild-type and vaccine strains of feline calicivirus
Abstract
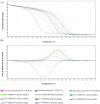
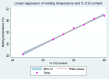
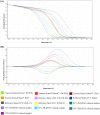
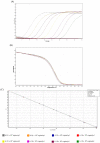
High-resolution melting (HRM) analysis, a post-polymerase chain reaction (PCR) application in a single closed tube, is the straightforward method for simultaneous detection, genotyping, and mutation scanning, enabling more significant dynamic detection and sequencing-free turnaround time. This study aimed to establish a combined reverse-transcription quantitative PCR and HRM (RT-qPCR-HRM) assay for diagnosing and genotyping feline calicivirus (FCV). This developed method was validated with constructed FCV plasmids, clinical swab samples from living cats, fresh-frozen lung tissues from necropsied cats, and four available FCV vaccines. We performed RT-qPCR to amplify a 99-base pair sequence, targeting a segment between open reading frame (ORF) 1 and ORF2. Subsequently, the HRM assay was promptly applied using Rotor-Gene Q® Software. The results significantly revealed simultaneous detection and genetic discrimination between commercially available FCV vaccine strains, wild-type Thai FCV strains, and VS-FCV strains within a single PCR reaction. There was no cross-reactivity with other feline common viruses, including feline herpesvirus-1, feline coronavirus, feline leukemia virus, feline immunodeficiency virus, and feline morbillivirus. The detection limit of the assay was 6.18 × 101 copies/µl. This study, therefore, is the first demonstration of the uses and benefits of the RT-qPCR-HRM assay for FCV detection and strain differentiation in naturally infected cats.
Keywords: High-resolution melting analysis; cat; feline calicivirus; polymerase chain reaction.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
The quadruplex TaqMan MGB fluorescent quantitative PCR method for simultaneous detection of feline panleukopenia virus, feline herpesvirus 1, feline calicivirus and feline infectious peritonitis virus.Front Cell Infect Microbiol. 2025 May 30;15:1581946. doi: 10.3389/fcimb.2025.1581946. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40521026 Free PMC article.
-
Detection and strain differentiation of feline calicivirus in conjunctival swabs by RT-PCR of the hypervariable region of the capsid protein gene.Arch Virol. 1998;143(7):1321-34. doi: 10.1007/s007050050378. Arch Virol. 1998. PMID: 9722877
-
Development of a novel reverse transcription PCR and its application to field sample testing for feline calicivirus prevalence in healthy stray cats in Korea.J Vet Sci. 2020 Sep;21(5):e71. doi: 10.4142/jvs.2020.21.e71. J Vet Sci. 2020. PMID: 33016018 Free PMC article.
-
Feline calicivirus.Vet Res. 2007 Mar-Apr;38(2):319-35. doi: 10.1051/vetres:2006056. Epub 2007 Feb 13. Vet Res. 2007. PMID: 17296159 Review.
-
An Update on Feline Calicivirus.Schweiz Arch Tierheilkd. 2022 Mar;164(3):225-241. doi: 10.17236/sat00346. Schweiz Arch Tierheilkd. 2022. PMID: 35232714 Review. English.
Cited by
-
Natural fatal infection of Tembusu virus in bottlenose dolphins in Thailand.Sci Rep. 2025 Mar 22;15(1):9917. doi: 10.1038/s41598-025-93477-5. Sci Rep. 2025. PMID: 40121312 Free PMC article.
-
Update on feline calicivirus: viral evolution, pathogenesis, epidemiology, prevention and control.Front Microbiol. 2024 May 2;15:1388420. doi: 10.3389/fmicb.2024.1388420. eCollection 2024. Front Microbiol. 2024. PMID: 38756726 Free PMC article. Review.
-
Salivary peptidomic profiling of chronic gingivostomatitis in cats by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry and nanoscale liquid chromatography-tandem mass spectrometry.J Vet Intern Med. 2025 Jan-Feb;39(1):e17247. doi: 10.1111/jvim.17247. Epub 2024 Nov 22. J Vet Intern Med. 2025. PMID: 39576047 Free PMC article.
-
Comparison of PCR, Nested PCR, and RT-LAMP for Rapid Detection of Feline Calicivirus Infection in Clinical Samples.Animals (Basel). 2024 Aug 22;14(16):2432. doi: 10.3390/ani14162432. Animals (Basel). 2024. PMID: 39199965 Free PMC article.
-
Rapid Detection of Feline Calicivirus Using Lateral Flow Dipsticks Based on CRISPR/Cas13a System.Animals (Basel). 2024 Dec 18;14(24):3663. doi: 10.3390/ani14243663. Animals (Basel). 2024. PMID: 39765567 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
