Human thymic putative CD8αα precursors exhibit a biased TCR repertoire in single cell AIRR-seq
- PMID: 37853083
- PMCID: PMC10584817
- DOI: 10.1038/s41598-023-44693-4
Human thymic putative CD8αα precursors exhibit a biased TCR repertoire in single cell AIRR-seq
Abstract
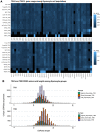
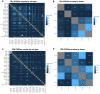
Thymic T cell development comprises T cell receptor (TCR) recombination and assessment of TCR avidity towards self-peptide-MHC complexes presented by antigen-presenting cells. Self-reactivity may lead to negative selection, or to agonist selection and differentiation into unconventional lineages such as regulatory T cells and CD8[Formula: see text] T cells. To explore the effect of the adaptive immune receptor repertoire on thymocyte developmental decisions, we performed single cell adaptive immune receptor repertoire sequencing (scAIRR-seq) of thymocytes from human young paediatric thymi and blood. Thymic PDCD1+ cells, a putative CD8[Formula: see text] T cell precursor population, exhibited several TCR features previously associated with thymic and peripheral ZNF683+ CD8[Formula: see text] T cells, including enrichment of large and positively charged complementarity-determining region 3 (CDR3) amino acids. Thus, the TCR repertoire may partially explain the decision between conventional vs. agonist selected thymocyte differentiation, an aspect of importance for the development of therapies for patients with immune-mediated diseases.
© 2023. Springer Nature Limited.
Conflict of interest statement
V.G. declares advisory board positions in aiNET GmbH, Enpicom B.V, Absci, Omniscope, and Diagonal Therapeutics. V.G. is a consultant for Adaptyv Biosystems, Specifica Inc, Roche/Genentech, immunai, and LabGenius. Remaining authors declare that they have no competing interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

