Clinical trial links oncolytic immunoactivation to survival in glioblastoma
- PMID: 37853118
- PMCID: PMC10620094
- DOI: 10.1038/s41586-023-06623-2
Clinical trial links oncolytic immunoactivation to survival in glioblastoma
Abstract
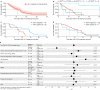
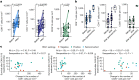
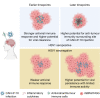
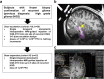
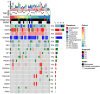
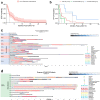
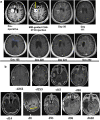
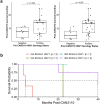
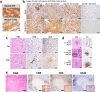
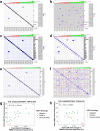
Immunotherapy failures can result from the highly suppressive tumour microenvironment that characterizes aggressive forms of cancer such as recurrent glioblastoma (rGBM)1,2. Here we report the results of a first-in-human phase I trial in 41 patients with rGBM who were injected with CAN-3110-an oncolytic herpes virus (oHSV)3. In contrast to other clinical oHSVs, CAN-3110 retains the viral neurovirulence ICP34.5 gene transcribed by a nestin promoter; nestin is overexpressed in GBM and other invasive tumours, but not in the adult brain or healthy differentiated tissue4. These modifications confer CAN-3110 with preferential tumour replication. No dose-limiting toxicities were encountered. Positive HSV1 serology was significantly associated with both improved survival and clearance of CAN-3110 from injected tumours. Survival after treatment, particularly in individuals seropositive for HSV1, was significantly associated with (1) changes in tumour/PBMC T cell counts and clonal diversity, (2) peripheral expansion/contraction of specific T cell clonotypes; and (3) tumour transcriptomic signatures of immune activation. These results provide human validation that intralesional oHSV treatment enhances anticancer immune responses even in immunosuppressive tumour microenvironments, particularly in individuals with cognate serology to the injected virus. This provides a biological rationale for use of this oncolytic modality in cancers that are otherwise unresponsive to immunotherapy (ClinicalTrials.gov: NCT03152318 ).
© 2023. The Author(s).
Conflict of interest statement
J.D.B. has an equity position in Treovir, an oHSV clinical stage company and is a member of the POCKiT Diagnostics board of scientific advisors; and has a provisional patent (application number 63/273,577) entitled ‘Methods and formulations related to the intrathecal delivery of oncolytic viruses’. W.L.B. consulted for Stryker. P.P. is cofounder and member of the board of directors of Ternalys Therapeutics; he is also named as an inventor on patents related to non-coding RNA technology. P.Y.W. received research support from AstraZeneca/Medimmune, Beigene, Celgene, Chimerix, Eli Lily, Genentech/Roche, Kazia, MediciNova, Merck, Novartis, Nuvation Bio, Puma, Servier, Vascular Biogenics and VBI Vaccines and served on advisory boards for AstraZeneca, Bayer, Black Diamond, Boehringer Ingelheim, Boston Pharmaceuticals, Celularity, Chimerix, Genenta, GlaxoSmithKline, Karyopharm, Merck, Mundipharma, Novartis, Novocure, Nuvation Bio, Prelude Therapeutics, Sapience, Servier, Sagimet, Vascular Biogenics and VBI Vaccines, and on data safety monitoring committees for Day One Bio and Novocure. L.N. serves as a consultant for Ono and Brave Bio. L.N.G.C. received research support from Merck (to the Dana-Farber Cancer Institute); he also has received research support from the NIH, the American Society of Clinical Oncology and the Robert Wood-Johnson Foundation. E.Q.L. receives royalties from Wolter Kluwer (Up to Date) and consulting fees from GCAR. T.B. receives clinical trial support from ONO Pharmaceuticals, publishing royalties from UpToDate and Oxford University Press. S.J.R. receives research support from Bristol Myers-Squibb and KITE/Gilead; and is on the scientific advisory board for Immunitas Therapeutics. K.K. is employed by and owns equity in Clearpoint. E. Tikhonova, N. Miheecheva, D. Tabakov, N. Shin, A.G., A. Shumskiy and F.F. are employed by BostonGene and have equity options in BostonGene. E.A.-C. is a founder, board member of and holds equity in Candel Therapeutics. L.K.A. is co-founder and holds equity in Candel Therapeutics. D. Krisky, J.W., A.M., C.M., P.P.T. and F.B. are employees of and hold equity in Candel Therapeutics. I.T. is an advisory board member of Immunitas Therapeutics. M.L.S. and K.W.W. are equity holders, scientific co-founders and advisory board members of Immunitas Therapeutics. D.A.R. is an advisor to Agios, AnHeart Therapeutics, Avita Biomedical, Blue Rock Therapeutics, Bristol Myers Squibb, Boston Biomedical, CureVac, Del Mar Pharma, DNAtrix, Hoffman-LaRoche, Imvax, Janssen, Kiyatec, Medicenna Therapeutics, Neuvogen, Novartis, Novocure, Pyramid, Sumitomo Dainippon Pharma, Vivacitas Oncology and Y-mabs Therapeutics. E.A.C. is an advisor to Amacathera, Bionaut Labs, Genenta, Insightec, DNAtrix, Seneca Therapeutics and Theravir; he has equity options in Bionaut Laboratories, DNAtrix, Immunomic Therapeutics, Seneca Therapeutics and Ternalys Therapeutics; he is co-founder and on the board of directors of Ternalys Therapeutics. Patents related to oHSV and CAN-3110 are under the possession of Brigham and Women’s Hospital with E.A.C. and H.N. named as co-inventors. These patents have been licensed to Candel Therapeutics. Present and future milestone license fees and future royalty fees are distributed to Brigham and Women’s Hospital from Candel. The other authors declare no competing interests.
Figures









Comment in
-
Glioblastoma tackled with oncolytic virus.Nat Rev Neurol. 2023 Dec;19(12):711. doi: 10.1038/s41582-023-00903-1. Nat Rev Neurol. 2023. PMID: 37940647 No abstract available.
References
-
- Chiocca, E. A., Nakashima, H., Kasai, K., Fernandez, S. A. & Oglesbee, M. Preclinical toxicology of rQNestin34.5v.2: an oncolytic herpes virus with transcriptional regulation of the ICP34.5 neurovirulence gene. Mol. Ther. Methods Clin. Dev.17, 871–893 (2020). 10.1016/j.omtm.2020.03.028 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

