Bioinformatics-led discovery of ferroptosis-associated diagnostic biomarkers and molecule subtypes for tuberculosis patients
- PMID: 37853432
- PMCID: PMC10585777
- DOI: 10.1186/s40001-023-01371-5
Bioinformatics-led discovery of ferroptosis-associated diagnostic biomarkers and molecule subtypes for tuberculosis patients
Abstract
Background: Ferroptosis is closely associated with the pathophysiological processes of many diseases, such as infection, and is characterized by the accumulation of excess lipid peroxides on the cell membranes. However, studies on the ferroptosis-related diagnostic markers in tuberculosis (TB) is still lacking. Our study aimed to explore the role of ferroptosis-related biomarkers and molecular subtypes in TB.
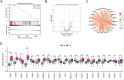
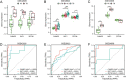
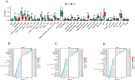
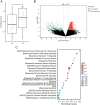
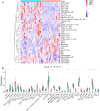
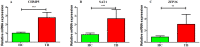
Methods: GSE83456 dataset was applied to identify ferroptosis-related genes (FRGs) associated with TB, and GSE42826, GSE28623, and GSE34608 datasets for external validation of core biomarkers. Core FRGs were identified using weighted gene co-expression network analysis (WGCNA). Subsequently, two ferroptosis-related subtypes were constructed based on ferroptosis score, and differently expressed analysis, GSEA, GSEA, immune cell infiltration analysis between the two subtypes were performed.Affiliations: Please check and confirm that the authors and their respective affiliations have been correctly identified and amend if necessary.correctly RESULTS: A total of 22 FRGs were identified, of which three genes (CHMP5, SAT1, ZFP36) were identified as diagnostic biomarkers that were enriched in pathways related to immune-inflammatory response. In addition, TB patients were divided into high- and low-ferroptosis subtypes (HF and LF) based on ferroptosis score. HF patients had activated immune- and inflammation-related pathways and higher immune cell infiltration levels than LF patients.
Conclusion: Three potential diagnostic biomarkers and two ferroptosis-related subtypes were identified in TB patients, which would help to understand the pathogenesis of TB.Author names: Kindly check and confirm the process of the author names [2,4]correctly.
Keywords: Diagnostic biomarkers; Ferroptosis; Molecular types; Tuberculosis; xCell.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
All authors declared that there was no competing interests.
Figures



Similar articles
-
Assessment of ferroptosis-associated gene signatures as potential biomarkers for differentiating latent from active tuberculosis in children.Microb Genom. 2023 May;9(5):mgen000997. doi: 10.1099/mgen.0.000997. Microb Genom. 2023. PMID: 37163321 Free PMC article.
-
Exploration of ferroptosis and necroptosis-related genes and potential molecular mechanisms in psoriasis and atherosclerosis.Front Immunol. 2024 Jul 12;15:1372303. doi: 10.3389/fimmu.2024.1372303. eCollection 2024. Front Immunol. 2024. PMID: 39072329 Free PMC article.
-
Establishment of Ferroptosis-Associated Molecular Subtypes and Hub Genes Related to the Immune Microenvironment of Heart Failure.Front Biosci (Landmark Ed). 2023 Oct 19;28(10):246. doi: 10.31083/j.fbl2810246. Front Biosci (Landmark Ed). 2023. PMID: 37919056
-
Mycobacterium tuberculosis infection induces a novel type of cell death: Ferroptosis.Biomed Pharmacother. 2024 Aug;177:117030. doi: 10.1016/j.biopha.2024.117030. Epub 2024 Jun 24. Biomed Pharmacother. 2024. PMID: 38917759 Review.
-
Ferroptosis-Associated Classifier and Indicator for Prognostic Prediction in Cutaneous Melanoma.J Oncol. 2021 Oct 28;2021:3658196. doi: 10.1155/2021/3658196. eCollection 2021. J Oncol. 2021. PMID: 34745259 Free PMC article. Review.
Cited by
-
Distinguish active tuberculosis with an immune-related signature and molecule subtypes: a multi-cohort analysis.Sci Rep. 2024 Nov 28;14(1):29564. doi: 10.1038/s41598-024-80072-3. Sci Rep. 2024. PMID: 39609541 Free PMC article.
-
The Role of Ferroptosis and Cuproptosis in Tuberculosis Pathogenesis: Implications for Therapeutic Strategies.Curr Issues Mol Biol. 2025 Feb 5;47(2):99. doi: 10.3390/cimb47020099. Curr Issues Mol Biol. 2025. PMID: 39996820 Free PMC article. Review.
-
Bioinformatics-Driven Identification of Ferroptosis-Related Gene Signatures Distinguishing Active and Latent Tuberculosis.Genes (Basel). 2025 Jun 18;16(6):716. doi: 10.3390/genes16060716. Genes (Basel). 2025. PMID: 40565608 Free PMC article.
-
Identification of ferroptosis-related gene signature for tuberculosis diagnosis and therapy efficacy.iScience. 2024 Jun 4;27(7):110182. doi: 10.1016/j.isci.2024.110182. eCollection 2024 Jul 19. iScience. 2024. PMID: 38989455 Free PMC article.
-
Identification of diagnostic biomarkers and molecular subtype analysis associated with m6A in Tuberculosis immunopathology using machine learning.Sci Rep. 2024 Dec 2;14(1):29982. doi: 10.1038/s41598-024-81790-4. Sci Rep. 2024. PMID: 39622968 Free PMC article.
References
-
- Furin J, Cox H, Pai M. Tuberculosis. Lancet. 2019;393:1642–1656. - PubMed
-
- Harding E. WHO global progress report on tuberculosis elimination. Lancet Respir Med. 2020;8:19. - PubMed
-
- Torres-Juarez F, Trejo-Martínez LA, Layseca-Espinosa E, Leon-Contreras JC, Enciso-Moreno JA, Hernandez-Pando R, Rivas-Santiago B. Platelets immune response against Mycobacterium tuberculosis infection. Microb Pathog. 2021;153:104768. - PubMed
-
- Natarajan A, Beena PM, Devnikar AV, Mali S. A systemic review on tuberculosis. Indian J Tuberc. 2020;67:295–311. - PubMed
-
- Young DB, Gideon HP, Wilkinson RJ. Eliminating latent tuberculosis. Trends Microbiol. 2009;17:183–188. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

