Multiscale simulations reveal TDP-43 molecular-level interactions driving condensation
- PMID: 37853696
- PMCID: PMC10720261
- DOI: 10.1016/j.bpj.2023.10.016
Multiscale simulations reveal TDP-43 molecular-level interactions driving condensation
Abstract
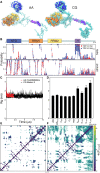
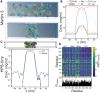
The RNA-binding protein TDP-43 is associated with mRNA processing and transport from the nucleus to the cytoplasm. TDP-43 localizes in the nucleus as well as accumulating in cytoplasmic condensates such as stress granules. Aggregation and formation of amyloid-like fibrils of cytoplasmic TDP-43 are hallmarks of numerous neurodegenerative diseases, most strikingly present in >90% of amyotrophic lateral sclerosis (ALS) patients. If excessive accumulation of cytoplasmic TDP-43 causes, or is caused by, neurodegeneration is presently not known. In this work, we use molecular dynamics simulations at multiple resolutions to explore TDP-43 self- and cross-interaction dynamics. A full-length molecular model of TDP-43, all 414 amino acids, was constructed from select structures of the protein functional domains (N-terminal domain, and two RNA recognition motifs, RRM1 and RRM2) and modeling of disordered connecting loops and the low complexity glycine-rich C-terminus domain. All-atom CHARMM36m simulations of single TDP-43 proteins served as guides to construct a coarse-grained Martini 3 model of TDP-43. The Martini model and a coarser implicit solvent C⍺ model, optimized for disordered proteins, were subsequently used to probe TDP-43 interactions; self-interactions from single-chain full-length TDP-43 simulations, cross-interactions from simulations with two proteins and simulations with assemblies of dozens to hundreds of proteins. Our findings illustrate the utility of different modeling scales for accessing TDP-43 molecular-level interactions and suggest that TDP-43 has numerous interaction preferences or patterns, exhibiting an overall strong, but dynamic, association and driving the formation of biomolecular condensates.
Copyright © 2023 Biophysical Society. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

