The differential prognostic implications of PD-L1 expression in the outcomes of Filipinos with EGFR-mutant NSCLC treated with tyrosine kinase inhibitors
- PMID: 37854154
- PMCID: PMC10579834
- DOI: 10.21037/tlcr-23-118
The differential prognostic implications of PD-L1 expression in the outcomes of Filipinos with EGFR-mutant NSCLC treated with tyrosine kinase inhibitors
Abstract
Background: The tumor immune microenvironment influences tumor evolution in non-small cell lung cancer (NSCLC). Yet, the prognostic value of programmed death-ligand 1 (PD-L1) in epidermal growth factor receptor (EGFR)-mutant NSCLC remains controversial. Additionally, prognostic studies in Filipinos with EGFR-mutant NSCLC remain unexplored to this day.
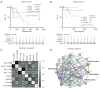
Methods: We prospectively studied the outcomes of EGFR-mutant NSCLC in Filipino cohort, and retrospectively verified the survival trend using The Cancer Genome Atlas (TCGA) cohort. Kaplan-Meier method and generalized linear regression were used to assess survival. Expression and DNA methylation of cluster of differentiation 274 (CD274, gene that codes for PD-L1) were examined from TCGA tumor profiles. Pearson's correlation was used to correlate PD-L1 expression with outcomes associated with occurrence of EGFR mutations, tyrosine kinase inhibitor (TKI) types, and programmed cell death protein 1 (PD-1) expression. Proteome network analysis was used to examine the correlation between drug resistance and PD-L1.
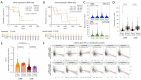
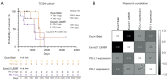
Results: PD-L1 positivity was associated with significantly longer progression-free survival (PFS; P=0.0096) but had a significantly contrasting influence in the overall survival (OS; P=0.0011). PD-L1 positivity (in both protein and RNA) was associated with longer median OS (mOS) in exon21 L858R, whereas, negativity was associated with longer mOS in exon19 deletion (exon19del). Stratification (high, low, negative) of PD-L1 expression lacked significant prognostic value (all P>0.05). PD-L1/CD274 expression (P<0.05) and DNA methylation (P<0.001) vary significantly among NSCLC subtypes and in different disease stages. Erlotinib treatment produced the longest median progression-free survival (mPFS; 874 days) relative to other EGFR-TKIs (137-311 days). PD-L1 lacked a significant correlation with EGFR-TKIs. Consistent with the immune-regulation activities of PD-1, higher expression leads to relatively shorter mOS. PD-1 correlated positively with PD-L1 expression and occurrence of exon21 L858R.
Conclusions: PD-L1 differentially influenced the outcomes of Filipinos with EGFR-mutant NSCLC. NSCLC subtypes, disease stage, and PD-1 expression may impact the collective outcomes associated with PD-L1 and EGFR-sensitizing mutations.
Keywords: EGFR tyrosine kinase inhibitor (EGFR-TKI); Filipinos NSCLC; Programmed death ligand-1 (PD-L1); epidermal growth factor receptor mutations (EGFR mutations); non-small cell lung cancer (NSCLC).
2023 Translational Lung Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-23-118/coif). The authors have no conflicts of interest to declare.
Figures


References
-
- WHO-GLOBOCAN. World Health Organization International Agency for Re-search on Cancer. 2020 Available online: https://gco.iarc.fr/today/data/factsheets/cancers/39-All-cancers-fact-sh... (accessed July 26, 2022).
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
