The Largest Chinese Cohort Study Indicates Homologous Recombination Pathway Gene Mutations as Another Major Genetic Risk Factor for Colorectal Cancer with Heterogeneous Clinical Phenotypes
- PMID: 37854294
- PMCID: PMC10581333
- DOI: 10.34133/research.0249
The Largest Chinese Cohort Study Indicates Homologous Recombination Pathway Gene Mutations as Another Major Genetic Risk Factor for Colorectal Cancer with Heterogeneous Clinical Phenotypes
Abstract
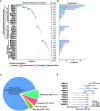
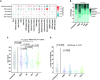
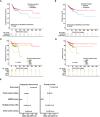
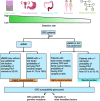
While genetic factors were associated with over 30% of colorectal cancer (CRC) patients, mutations in CRC-susceptibility genes were identified in only 5% to 10% of these patients. Besides, previous studies on hereditary CRC were largely designed to analyze germline mutations in patients with single genetic high-risk factor, which limited understanding of the association between genotype and phenotypes. From January 2015 to December 2018, we retrospectively enrolled 2,181 patients from 8,270 consecutive CRC cases, covering 5 categories of genetic high-risk factors. Leukocyte genomic DNA was analyzed for germline mutations in cancer predisposition genes. The germline mutations under each category were detected and analyzed in association with CRC susceptibility, clinical phenotypes, and prognoses. A total of 462 pathogenic variants were detected in 19.3% of enrolled CRC patients. Mismatch repair gene mutation was identified in 9.1% of patients, most prevalent across all high-risk groups. Homologous recombination (HR) gene mutations were detected in 6.5% of cases, penetrated in early-onset and extra-colonic cancer risk groups. Mutations in HR genes, including BARD1, RAD50, and ATM, were found to increase CRC risk with odds ratios of 2.8-, 3.1-, and 3.1-fold, respectively. CRC patients with distinct germline mutations manifested heterogeneous phenotypes in clinicopathology and long-term prognoses. Thus, germline mutation screenings should be performed for CRC patients with any of those genetic risk factors. This study also reveals that HR gene mutations may be another major driver for increased CRC risk.
Copyright © 2023 Yun Xu et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures


References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. - PubMed
-
- GBD 2017 Colorectal Cancer Collaborators. The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2019;4(12):913–933. - PMC - PubMed
-
- Yang Y, Han Z, Li X, Huang A, Shi J, Gu J, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Center of Gastrointestinal Surgery, Peking University Cancer Hospital & Institute, Beijing 100142, China; Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Gastrointestinal Oncology, Peking University Cancer Hospital & Institute, Beijing 100142, China; Peking-Tsinghua Center for Life Science, Peking University International Cancer Center, Beijing 100142, China; Department of Gastrointestinal Surgery, Peking University Shougang Hospital, Beijing 100144, China. Epidemiology and risk factors of colorectal cancer in China. Chin J Cancer Res. 2020;32(6):729–741. - PMC - PubMed
-
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. - PubMed
-
- Monahan KJ, Bradshaw N, Dolwani S, Desouza B, Dunlop MG, East JE, Ilyas M, Kaur A, Lalloo F, Latchford A, et al. . Guidelines for the management of hereditary colorectal cancer from the British Society of Gastroenterology (BSG)/Association of Coloproctology of Great Britain and Ireland (ACPGBI)/United Kingdom cancer genetics group (UKCGG). Gut. 2020;69(3):411–444. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

