Liver injury monitoring using dynamic fluorescence molecular tomography based on a time-energy difference strategy
- PMID: 37854546
- PMCID: PMC10581805
- DOI: 10.1364/BOE.498092
Liver injury monitoring using dynamic fluorescence molecular tomography based on a time-energy difference strategy
Abstract
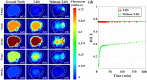
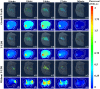
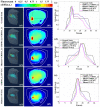
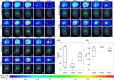
Dynamic fluorescence molecular tomography (DFMT) is a promising molecular imaging technique that offers the potential to monitor fast kinetic behaviors within small animals in three dimensions. Early monitoring of liver disease requires the ability to distinguish and analyze normal and injured liver tissues. However, the inherent ill-posed nature of the problem and energy signal interference between the normal and injured liver regions limit the practical application of liver injury monitoring. In this study, we propose a novel strategy based on time and energy, leveraging the temporal correlation in fluorescence molecular imaging (FMI) sequences and the metabolic differences between normal and injured liver tissue. Additionally, considering fluorescence signal distribution disparity between the injured and normal regions, we designed a universal Golden Ratio Primal-Dual Algorithm (GRPDA) to reconstruct both the normal and injured liver regions. Numerical simulation and in vivo experiment results demonstrate that the proposed strategy can effectively avoid signal interference between liver and liver injury energy and lead to significant improvements in morphology recovery and positioning accuracy compared to existing approaches. Our research presents a new perspective on distinguishing normal and injured liver tissues for early liver injury monitoring.
© 2023 Optica Publishing Group under the terms of the Optica Open Access Publishing Agreement.
Conflict of interest statement
The authors declare no potential conflict of interests.
Figures



References
-
- Horowitz J. M., Venkatesh S. K., Ehman R. L., Jhaveri K., Kamath P., Ohliger M. A., Samir A. E., Silva A. C., Taouli B., Torbenson M. S., Michael L. B., Miller H. F., “Evaluation of hepatic fibrosis: a review from the society of abdominal radiology disease focus panel,” Abdom. Radiol. 42(8), 2037–2053 (2017). 10.1007/s00261-017-1211-7 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
