In silico virtual screening for the identification of novel inhibitors against dihydrodipicolinate reductase (DapB) of Mycobacterium tuberculosis, a key enzyme of diaminopimelate pathway
- PMID: 37855602
- PMCID: PMC10714930
- DOI: 10.1128/spectrum.01359-23
In silico virtual screening for the identification of novel inhibitors against dihydrodipicolinate reductase (DapB) of Mycobacterium tuberculosis, a key enzyme of diaminopimelate pathway
Abstract
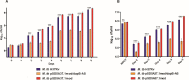
Non-compliance to lengthy antituberculosis (TB) treatment regimen, associated side effects, and emergence of drug-resistant strains of Mycobacterium tuberculosis (M. tb) emphasize the need to develop more effective anti-TB drugs. Here, we have evaluated the role of M. tb dihydrodipicolinate reductase (DapB), a component of the diaminopimelate pathway, which is involved in the biosynthesis of both lysine and mycobacterial cell wall. We showed that DapB is essential for the in vitro as well as intracellular growth of M. tb. We further utilized M. tb DapB, as a target for identification of inhibitors by employing in silico virtual screening, and conducted various in vitro screening assays to identify inhibitors with potential to inhibit DapB activity and in vitro and intracellular growth of M. tb with no significant cytotoxicity against various mammalian cell lines. Altogether, M. tb DapB serves as an important drug target and a hit molecule, namely, 4-(3-Phenylazoquinoxalin-2-yl) butanoic acid methyl ester has been identified as an antimycobacterial molecule in our study.
Keywords: Mycobacterium tuberculosis; tuberculosis; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Vilchèze C. 2020. Mycobacterial cell wall: a source of successful targets for old and new drugs. Applied Sciences 10:2278. doi:10.3390/app10072278 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

