CaeNDR, the Caenorhabditis Natural Diversity Resource
- PMID: 37855690
- PMCID: PMC10767927
- DOI: 10.1093/nar/gkad887
CaeNDR, the Caenorhabditis Natural Diversity Resource
Abstract
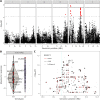
Studies of model organisms have provided important insights into how natural genetic differences shape trait variation. These discoveries are driven by the growing availability of genomes and the expansive experimental toolkits afforded to researchers using these species. For example, Caenorhabditis elegans is increasingly being used to identify and measure the effects of natural genetic variants on traits using quantitative genetics. Since 2016, the C. elegans Natural Diversity Resource (CeNDR) has facilitated many of these studies by providing an archive of wild strains, genome-wide sequence and variant data for each strain, and a genome-wide association (GWA) mapping portal for the C. elegans community. Here, we present an updated platform, the Caenorhabditis Natural Diversity Resource (CaeNDR), that enables quantitative genetics and genomics studies across the three Caenorhabditis species: C. elegans, C. briggsae and C. tropicalis. The CaeNDR platform hosts several databases that are continually updated by the addition of new strains, whole-genome sequence data and annotated variants. Additionally, CaeNDR provides new interactive tools to explore natural variation and enable GWA mappings. All CaeNDR data and tools are accessible through a freely available web portal located at caendr.org.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998; 282:2012–2018. - PubMed
-
- Chalfie M., Tu Y., Euskirchen G., Ward W.W., Prasher D.C.. Green fluorescent protein as a marker for gene expression. Science. 1994; 263:802–805. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

