DNA-Histone Cross-Link Formation via Hole Trapping in Nucleosome Core Particles
- PMID: 37856159
- PMCID: PMC10652223
- DOI: 10.1021/jacs.3c08135
DNA-Histone Cross-Link Formation via Hole Trapping in Nucleosome Core Particles
Abstract
Radical cations (holes) produced in DNA by ionizing radiation and other oxidants yield DNA-protein cross-links (DPCs). Detailed studies of DPC formation in chromatin via this process are lacking. We describe here a comprehensive examination of DPC formation within nucleosome core particles (NCPs), which are the monomeric component of chromatin. DNA holes are introduced at defined sites within NCPs that are constructed from the bottom-up. DPCs form at DNA holes in yields comparable to those of alkali-labile DNA lesions that result from water trapping. DPC-forming efficiency and site preference within the NCP are dependent on translational and rotational positioning. Mass spectrometry and the use of mutant histones reveal that lysine residues in histone N-terminal tails and amino termini are responsible for the DPC formation. These studies are corroborated by computational simulation at the microsecond time scale, showing a wide range of interactions that can precede DPC formation. Three consecutive dGs, which are pervasive in the human genome, including G-quadruplex-forming sequences, are sufficient to produce DPCs that could impact gene expression.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
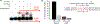





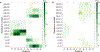
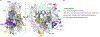
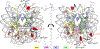

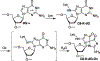
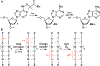

References
-
- Wei X; Peng Y; Bryan C; Yang K Mechanisms of DNA-Protein Cross-Link Formation and Repair. Biochim. Biophys. Acta 2021, 1869, 140669. - PubMed
-
- Nakano T; Mitsusada Y; Salem AMH; Shoulkamy MI; Sugimoto T; Hirayama R; Uzawa A; Furusawa Y; Ide H Induction of DNA-Protein Cross-Links by Ionizing Radiation and Their Elimination from the Genome. Mut. Res. 2015, 771, 45–50. - PubMed
-
- Vaz B; Popovic M; Newman JA; Fielden J; Aitkenhead H; Halder S; Singh AN; Vendrell I; Fischer R; Torrecilla I; Drobnitzky N; Freire R; Amor DJ; Lockhart PJ; Kessler BM; McKenna GW; Gileadi O; Ramadan K Metalloprotease Sprtn/Dvc1 Orchestrates Replication-Coupled DNA-Protein Crosslink Repair. Mol. Cell 2016, 64, 704–719. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

