Isolation, antimicrobial resistance and virulence characterization of Salmonella spp. from fresh foods in retail markets in Hangzhou, China
- PMID: 37856530
- PMCID: PMC10586686
- DOI: 10.1371/journal.pone.0292621
Isolation, antimicrobial resistance and virulence characterization of Salmonella spp. from fresh foods in retail markets in Hangzhou, China
Abstract
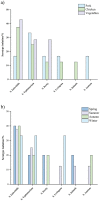
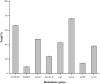
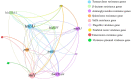
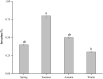
Salmonella can cause severe foodborne diseases. This study investigated the prevalence of Salmonella spp. in fresh foods in Hangzhou market and their harborage of antibiotic resistance and virulence genes, antibiotic susceptibility, and pathogenicity. A total of 500 samples (pork, n = 140; chicken, n = 128; vegetable, n = 232) were collected over a one-year period. Salmonella was found in 4.2% (21) of samples with the detection rate in pork, chicken and vegetables as 4.3% (6), 6.3% (8), and 3% (7), respectively. One Salmonella strain was recovered from each positive sample. The isolates were identified as six serotypes, of which S. Enteritidis (n = 7) and S. Typhimurium (n = 6) were the most predominant serotypes. The majority of isolates showed resistance to tetracycline (85.7%) and/or ciprofloxacin (71.4%). Tetracycline resistance genes showed the highest prevalence (90.5%). The occurrence of resistance genes for β-lactams (blaTEM-1, 66.7%; and blaSHV, 9.5%) and aminoglycosides (aadA1, 47.6%; Aac(3)-Ia, 19%) was higher than sulfonamides (sul1, 42.9%) and quinolones (parC, 38.1%). The virulence gene fimA was detected in 57.1% of isolates. Gene co-occurrence analysis implied that resistance genes were associated with virulence genes. Furthermore, selected S. Typhimurium isolates (n = 4) carrying different resistance and virulence genes up-regulated the secretions of cytokines IL-6 and IL-8 by Caco-2 cells in different degrees, suggesting that virulence genes may play a role in inflammatory transcription. In in vivo virulence test, microbiological counts in mouse feces and tissues showed that all included S. Typhimurium were able to infect mice, with one strain showing significantly higher virulence than others. In conclusion, this study indicates Salmonella contamination in fresh foods in Hangzhou market poses a risk to public health and it should be closely monitored to prevent and control foodborne diseases.
Copyright: © 2023 Qian et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Zx A, Min WA, Cw A, Cz A, Jl A, Gg B, et al. The emergence of extended-spectrum β-lactamase (ESBL)-producing Salmonella London isolates from human patients, retail meats and chickens in southern China and the evaluation of the potential risk factors of Salmonella London. Food Control. 2021;128. doi: 10.1016/j.foodcont.2021.108187 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

