Validating a Promoter Library for Application in Plasmid-Based Diatom Genetic Engineering
- PMID: 37857380
- PMCID: PMC10661051
- DOI: 10.1021/acssynbio.3c00163
Validating a Promoter Library for Application in Plasmid-Based Diatom Genetic Engineering
Abstract
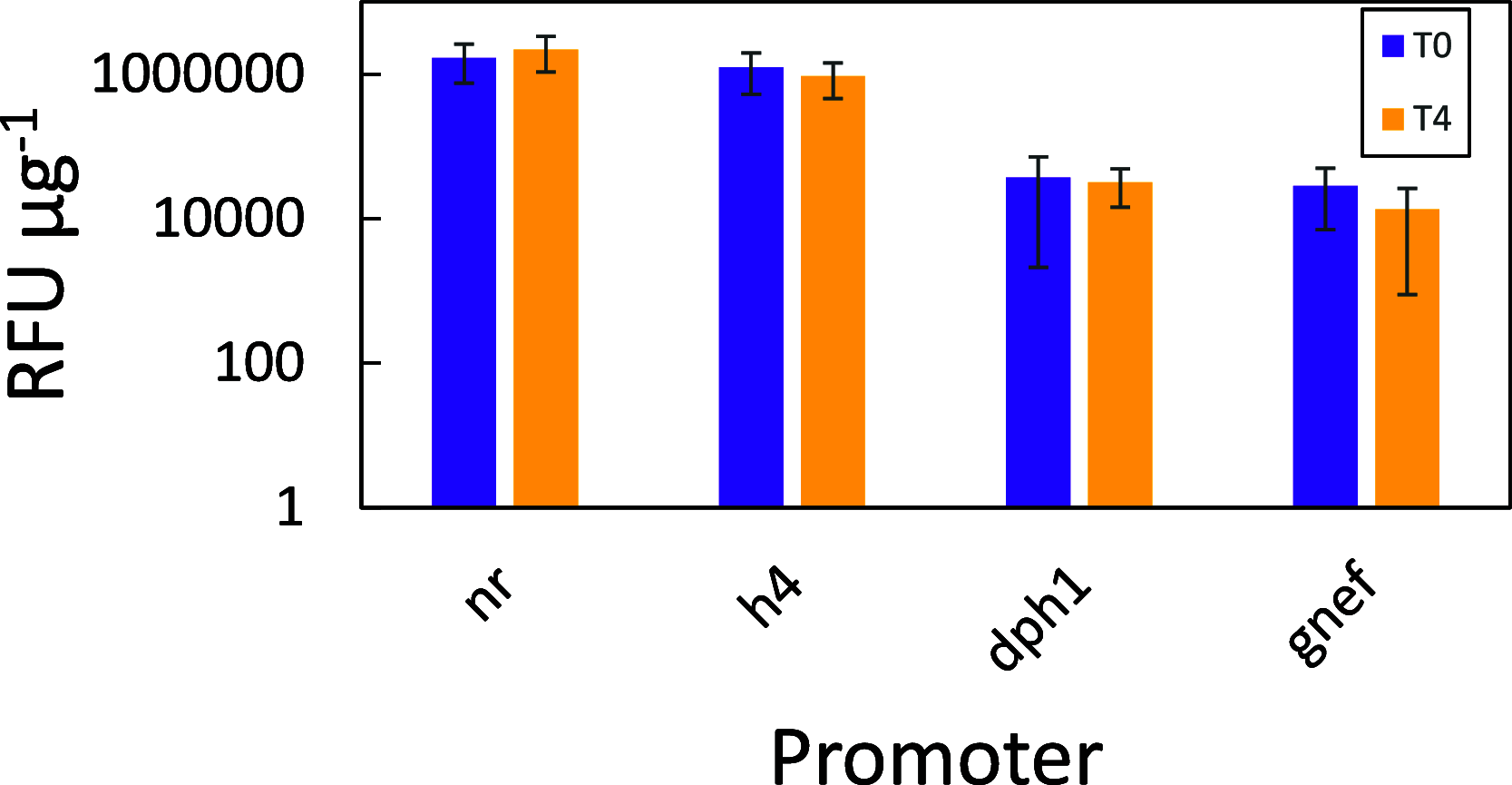
While diatoms are promising synthetic biology platforms, there currently exists a limited number of validated genetic regulatory parts available for genetic engineering. The standard method for diatom transformation, nonspecific introduction of DNA into chromosomes via biolistic particle bombardment, is low throughput and suffers from clonal variability and epigenetic effects. Recent developments in diatom engineering have demonstrated that autonomously replicating episomal plasmids serve as stable expression platforms for diverse gene expression technologies. These plasmids are delivered via bacterial conjugation and, when combined with modular DNA assembly technologies, provide a flexibility and speed not possible with biolistic-mediated strain generation. In order to expand the current toolbox for plasmid-based engineering in the diatom Phaeodactylum tricornutum, a conjugation-based forward genetics screen for promoter discovery was developed, and application to a diatom genomic DNA library defined 252 P. tricornutum promoter elements. From this library, 40 promoter/terminator pairs were delivered via conjugation on episomal plasmids, characterized in vivo, and ranked across 4 orders of magnitude difference in reporter gene expression levels.
Keywords: diatom; episomal gene expression; forward genetics; genetic engineering; parts registry; promoter characterization.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Behrenfeld M. J.; Halsey K. H.; Boss E.; Karp-Boss L.; Milligan A. J.; Peers G. Thoughts on the evolution and ecological niche of diatoms. Ecol. Monogr. 2021, 91, e0145710.1002/ecm.1457. - DOI
-
- Fabris M.; George J.; Kuzhiumparambil U.; Lawson C. A.; Jaramillo-Madrid A. C.; Abbriano R. M.; Vickers C. E.; Ralph P. Extrachromosomal Genetic Engineering of the Marine Diatom Phaeodactylum tricornutum Enables the Heterologous Production of Monoterpenoids. ACS Synth. Biol. 2020, 9, 598–612. 10.1021/acssynbio.9b00455. - DOI - PubMed

