YAEL: Your Advanced Electrode Localizer
- PMID: 37857509
- PMCID: PMC10591275
- DOI: 10.1523/ENEURO.0328-23.2023
YAEL: Your Advanced Electrode Localizer
Abstract
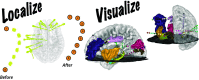
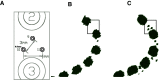
Intracranial electroencephalography (iEEG) provides a unique opportunity to record and stimulate neuronal populations in the human brain. A key step in neuroscience inference from iEEG is localizing the electrodes relative to individual subject anatomy and identified regions in brain atlases. We describe a new software tool, Your Advanced Electrode Localizer (YAEL), that provides an integrated solution for every step of the electrode localization process. YAEL is compatible with all common data formats to provide an easy-to-use, drop-in replacement for problematic existing workflows that require users to grapple with multiple programs and interfaces. YAEL's automatic extrapolation and interpolation functions speed localization, especially important in patients with many implanted stereotactic (sEEG) electrode shafts. The graphical user interface is presented in a web browser for broad compatibility and includes an interactive 3D viewer for easier localization of nearby sEEG contacts. After localization is complete, users may enter or import data into YAEL's 3D viewer to create publication-ready visualizations of electrodes and brain anatomy, including identified brain areas from atlases; the response to experimental tasks measured with iEEG; and clinical measures such as epileptiform activity or the results of electrical stimulation mapping. YAEL is free and open source and does not depend on any commercial software. Installation instructions for Mac, Windows, and Linux are available at https://yael.wiki.
Copyright © 2023 Wang et al.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
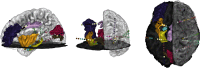



References
-
- Bartel F, Visser M, de Ruiter M, Belderbos J, Barkhof F, Vrenken H, de Munck JC, van Herk M; Alzheimer’s Disease Neuroimaging Initiative (2019) Non-linear registration improves statistical power to detect hippocampal atrophy in aging and dementia. Neuroimage Clin 23:101902. 10.1016/j.nicl.2019.101902 - DOI - PMC - PubMed
-
- Chang W, Cheng J, Allaire JJ, Sievert C, Schloerke B, Xie Y, Allen J, McPherson J, Dipert A, Borges B (2023) shiny: web application framework for R. Available at https://cran.r-project.org/web/packages/shiny/index.html.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
