Neuropeptide Y-Mediated Gut Microbiota Alterations Aggravate Postmenopausal Osteoporosis
- PMID: 37857552
- PMCID: PMC10667841
- DOI: 10.1002/advs.202303015
Neuropeptide Y-Mediated Gut Microbiota Alterations Aggravate Postmenopausal Osteoporosis
Abstract
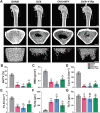
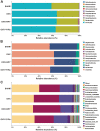
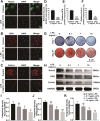
Postmenopausal osteoporosis (PMO) is often accompanied by neuroendocrine changes in the hypothalamus, which closely associates with the microbial diversity, community composition, and intestinal metabolites of gut microbiota (GM). With the emerging role of GM in bone metabolism, a potential neuroendocrine signal neuropeptide Y (NPY) mediated brain-gut-bone axis has come to light. Herein, it is reported that exogenous overexpression of NPY reduced bone formation, damaged bone microstructure, and up-regulated the expressions of pyroptosis-related proteins in subchondral cancellous bone in ovariectomized (OVX) rats, but Y1 receptor antagonist (Y1Ra) reversed these changes. In addition, it is found that exogenous overexpression of NPY aggravated colonic inflammation, impaired intestinal barrier integrity, enhanced intestinal permeability, and increased serum lipopolysaccharide (LPS) in OVX rats, and Y1Ra also reversed these changes. Most importantly, NPY and Y1Ra modulated the microbial diversity and changed the community composition of GM in OVX rats, and thereby affecting the metabolites of GM (e.g., LPS) entering the blood circulation. Moreover, fecal microbiota transplantation further testified the effect of NPY-mediated GM changes on bone. In vitro, LPS induced pyroptosis, reduced viability, and inhibited differentiation of osteoblasts. The study demonstrated the existence of NPY-mediated brain-gut-bone axis and it might be a novel emerging target to treat PMO.
Keywords: brain-gut-bone axis; gut microbiota; neuropeptide Y; osteoblast pyroptosis; postmenopausal osteoporosis.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
