Low mutation rate in epaulette sharks is consistent with a slow rate of evolution in sharks
- PMID: 37857613
- PMCID: PMC10587355
- DOI: 10.1038/s41467-023-42238-x
Low mutation rate in epaulette sharks is consistent with a slow rate of evolution in sharks
Abstract
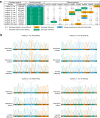
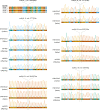
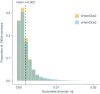
Sharks occupy diverse ecological niches and play critical roles in marine ecosystems, often acting as apex predators. They are considered a slow-evolving lineage and have been suggested to exhibit exceptionally low cancer rates. These two features could be explained by a low nuclear mutation rate. Here, we provide a direct estimate of the nuclear mutation rate in the epaulette shark (Hemiscyllium ocellatum). We generate a high-quality reference genome, and resequence the whole genomes of parents and nine offspring to detect de novo mutations. Using stringent criteria, we estimate a mutation rate of 7×10-10 per base pair, per generation. This represents one of the lowest directly estimated mutation rates for any vertebrate clade, indicating that this basal vertebrate group is indeed a slowly evolving lineage whose ability to restore genetic diversity following a sustained population bottleneck may be hampered by a low mutation rate.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Compagno LJV. Alternative life-history styles of cartilaginous fishes in time and space. Environ. Biol. Fishes. 1990;28:33–75. doi: 10.1007/BF00751027. - DOI
-
- Heithaus MR, Wirsing AJ, Dill LM. The ecological importance of intact top-predator populations: a synthesis of 15 years of research in a seagrass ecosystem. Mar. Freshw. Res. 2012;63:1039–1050. doi: 10.1071/MF12024. - DOI
-
- Stevens JD, Bonfil R, Dulvy NK, Walker PA. The effects of fishing on sharks, rays, and chimaeras (chondrichthyans), and the implications for marine ecosystems. ICES J. Mar. Sci. 2000;57:476–494. doi: 10.1006/jmsc.2000.0724. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

