RING finger protein 13 protects against nonalcoholic steatohepatitis by targeting STING-relayed signaling pathways
- PMID: 37857628
- PMCID: PMC10587083
- DOI: 10.1038/s41467-023-42420-1
RING finger protein 13 protects against nonalcoholic steatohepatitis by targeting STING-relayed signaling pathways
Abstract
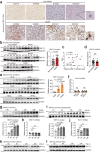
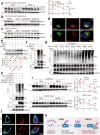
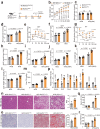
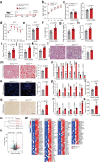
Nonalcoholic fatty liver disease (NAFLD) is the most common liver disorder worldwide. Recent studies show that innate immunity-related signaling pathways fuel NAFLD progression. This study aims to identify potent regulators of innate immunity during NAFLD progression. To this end, a phenotype-based high-content screening is performed, and RING finger protein 13 (RNF13) is identified as an effective inhibitor of lipid accumulation in vitro. In vivo gain- and loss-of-function assays are conducted to investigate the role of RNF13 in NAFLD. Transcriptome sequencing and immunoprecipitation-mass spectrometry are performed to explore the underlying mechanisms. We reveal that RNF13 protein is upregulated in the liver of individuals with NASH. Rnf13 knockout in hepatocytes exacerbate insulin resistance, steatosis, inflammation, cell injury and fibrosis in the liver of diet-induced mice, which can be alleviated by Rnf13 overexpression. Mechanically, RNF13 facilitates the proteasomal degradation of stimulator of interferon genes protein (STING) in a ubiquitination-dependent way. This study provides a promising innate immunity-related target for NAFLD treatment.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
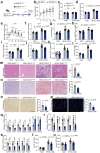
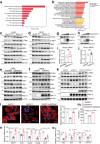
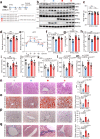
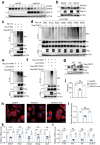

References
-
- Powell EE, Wong VW, Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397:2212–2224. - PubMed
-
- Loomba R, Friedman SL, Shulman GI. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell. 2021;184:2537–2564. - PubMed
-
- Arab JP, Arrese M, Trauner M. Recent insights into the pathogenesis of nonalcoholic fatty liver disease. Annu. Rev. Pathol. 2018;13:321–350. - PubMed
-
- Cai J, Xu M, Zhang X, Li H. Innate immune signaling in nonalcoholic fatty liver disease and cardiovascular diseases. Annu. Rev. Pathol. 2019;14:153–184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

